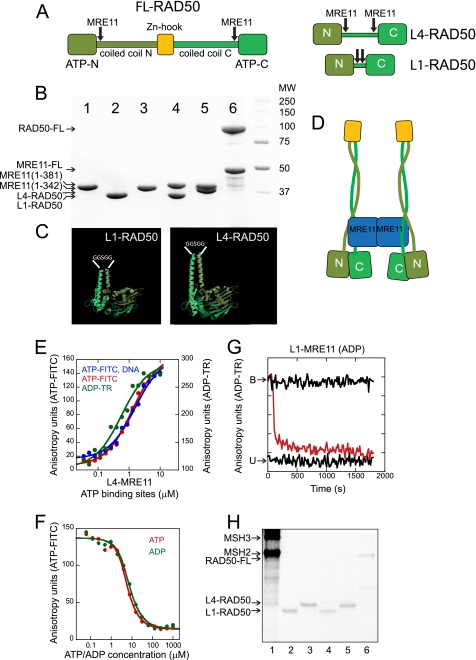
FIGURE 1.
Nucleotide binding to purified pfRAD50s and to their complexes with MRE11. A, linear representation of the FL pfRAD50 is shown. The MRE11 binding sites are indicated by the arrows, the zinc hook region is in gold, and the N- and C-coiled coil domains are dark and light green, respectively. The ATP binding sites (ATP N and C) are indicated. Right, linear representations of the truncated forms of RAD50 are shown. L4-RAD50 (L4) and L1-RAD50 (L1), each of which contains MRE11 binding sites, are shown. B, shown is SDS-PAGE resolution of the purified forms of pfRAD50 and pfMRE11 proteins used in the experiments. 1, MRE111–342; 2, L1-RAD50; 3, L4-RAD50; 4, L1-MRE111–381; 5, L4-MRE111–381; 6, FL-RAD50 and FL-MRE11. C, shown are pMOL images of the engineered L1-RAD50 and L4-RAD50 indicating the peptide linker (GGSGG) (white). D, shown is a schematic representation of the FL MRE11-RAD50 heterodimer. E, binding of BODIPY-labeled nucleotides for either ATP-FITC (red) or ADP-TR (green). Reactions containing DNA are indicated in blue. L4-MRE11 at the indicated protein concentration (expressed as concentration of ATP binding sites) was incubated at 25 °C for 5 min with 5 nm nucleotide in a 20-μl reaction volume. In the DNA binding curve (blue), the reactions contained 5 μm 40-bp double-stranded oligonucleotide. Anisotropy was measured on a TECAN plate reader using excitation and an emission wavelength of 470 and 522 nm for ATP-FITC and 590 and 615 nm for ADP-TR. Data were fitted (solid lines) to a Langmuir isotherm. F, in competition experiments, unlabeled nucleotide was added at the indicated concentrations to 3 μm L4-MRE11 (6 μm ATP binding sites) pre-bound with 5 nm ATP-FITC (red) or ADP-TR (green). G, displacement kinetics of prebound ADP-TR from pfRAD50 were measured using unlabeled ADP as a competitor. B indicates bound ADP-TR in the absence of nucleotide, and U indicates unbound pfRAD50. H, protein products of UV-cross-linking with labeled nucleotides is shown. 1, MSH2-MSH3 control; 2, L1; 3, L4; 4, L1-MRE11; 5, L4-MRE11; 6, FL. Reactions contained 100 nm concentrations of the indicated protein and 500 nm nucleotide. UV cross-linking of labeled nucleotides to RAD50 was performed in a Stratalinker 1800 cross-linker (Stratagene) for 7.5 min, and the products were resolved by SDS-PAGE.