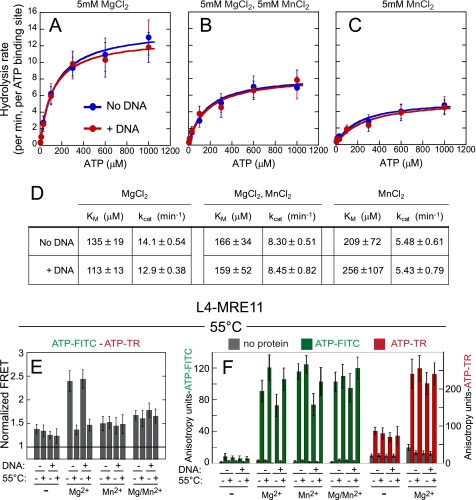
FIGURE 3.
ATP hydrolysis opens the RAD50-MRE11 complex. A–C, ATPase assays were performed at 55 °C. L4-MRE11 was added to nucleotides with (red) or without (blue) DNA (5 μm 40-bp double-stranded oligonucleotide) and incubated for 10 min at 55 °C. At 2, 4, 6, 10, and 20 min, 5-μl aliquots were removed, quenched, and analyzed by TLC. Data were fit (solid lines) to a Michaelis-Menten equation. Each reaction contained the indicated ion or combination of ions, 2 μm L4-MRE11 (4 μm ATP binding sites), and the indicated concentrations of [α-32P]ATP. D, the percentage of ADP formed was quantified by phosphorimaging and used to calculate ATP hydrolysis rates (expressed in turnover number per minute) in the presence or absence of DNA. E, shown is FRET between ATP-FITC and ATP-TR bound to L4-MRE11 at 55 °C in Mg2+, Mn2+, or a stoichiometric ratio of both Mg2+/Mn2+, or no ion was measured as described in Fig. 2. ATP hydrolysis and/or the addition of Mn2+ is sufficient to lose FRET and open the closed complex. F, shown is the nucleotide binding affinity measured at 55 °C for reactions in E using fluorescence anisotropy for ATP-FITC and ATP-TR. The excitation and emission of wavelengths were 470 and 522 nm for ATP-FITC and 590 and 615 nm for ATP-TR; gray is no protein, the green bar is ATP-FITC, and the red bar is ATP-TR. The presence (+) or absence (−) of DNA is indicated. Samples in the presence (+) or absence (−) of heating to 55 °C are indicated.