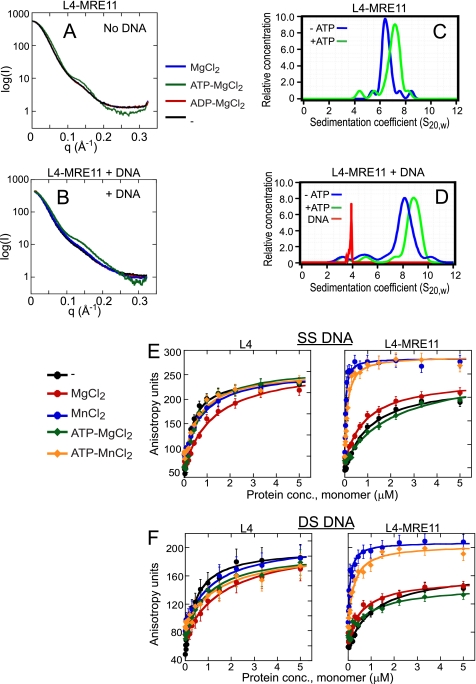
FIGURE 4.
DNA binding does not influence the conformation of L4 and L4-MRE11. A and B, SAXS of the L4-MRE11 heterotetramer (A) or the L4-MRE11 heterotetramer (B) in the presence of DNA in reactions containing (25 μm) protein and no ion (black), no nucleotide (blue), 200 μm ADP (Mg2+) (red), or 200 μm ATP (Mg2+) (green). The plot is of log intensity versus scattering vector (q)/Å. C and D, a plot of the relative protein concentration versus the sedimentation coefficient corrected for water at 20 °C (s20,w) of the L4-MRE11 heterotetramer (C) or the L4-MRE11 heterotetramer in the presence of DNA (D) and in the presence (green) or the absence (blue) of ATP (Mg2+). Red indicates the (s20,w) for 40-bp double-stranded DNA in the absence of protein. E and F, the open complex binds best to DNA. L4 or L4-MRE11 binding to a 2 nm 5′-FITC-labeled single strand of 40 nucleotides (supplemental sequences, 40 bot-FITC) or double-stranded DNA of 40 base pairs (supplemental sequences, 40 bot and 40 top) was measured by fluorescence anisotropy. Shown is anisotropy versus protein concentration. DNA was incubated with the indicated amount of protein (expressed as concentration of ATP binding sites) for 5 min at 25 °C in a 20-μl reaction volume. The samples contained 5 mm MgCl2 or MnCl2 and 200 μm ATP as indicated. Anisotropy was measured in a TECAN plate reader using excitation and emission wavelengths of 470 and 522 nm, respectively. Data were fit (solid lines) to the Langmuir isotherm.