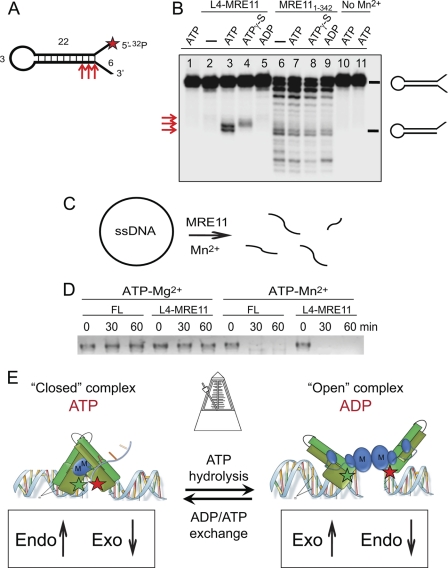
FIGURE 6.
ATP binding stimulates endonuclease activity of the RAD50-MRE11 complex. A, shown is a schematic representation of the DNA hairpin substrate used in the endonuclease assays. The asterisk marks position of the 32P label. The numbers indicate the nucleotide length in each segment of the template. B, assays contained 2 μm MRE11 or L4-MRE11 protein, 200 μm nucleotide, 5 mm concentrations of both MgCl2 and MnCl2, and ∼1 nm DNA substrate (10,000 cpm). Reactions were incubated at 55 °C for 2 h and stopped with gel loading buffer, and the products were resolved by 12% urea-PAGE followed by phosphorimaging. Lane 1, templates with ATP but no L4-MRE11; lane 2, L4-MRE11 with no nucleotide; lane 3, L4-MRE11 and ATP; lane 4, L4-MRE11 and ATPγS; lane 5, L4-MRE11 and ADP; lane 6, MRE111–342 and no nucleotide; lanes 7–9, MRE111–342 with ATP, ATPγS, and ADP, respectively; lane 10, ATP and L4-MRE11 and no manganese; lane 11, MRE111–342 and ATP no manganese. Migration of the endonuclease products is shown on the right. Red arrows (left) are reaction products corresponding to A. C, shown is an endonuclease assay using plasmid DNA. The assays contained 2 μg of ss plasmid, 1 μm FL or L4-MRE11 protein, and 1 mm ATP incubated with 5 mm concentrations of the indicated cation. Both full-length RAD50-MRE11 (FL) and L4-MRE11 were used, as indicated. Samples were taken at 0, 30, and 60 min. Disappearance of the band indicates incision. E, shown is a switching model for RAD50-MRE11. Butterfly-like opening and closing act as a metronome for switching the nucleolytic activity of MRE11 between exonuclease and endonuclease functions. Nucleotide exchange resets the clock.