Abstract
Two new marine-derived sesquiterpene benzoquinones which we designate as neopetrosiquinone A (1) and B (2), have been isolated from a deep-water sponge of the family Petrosiidae. The structures were elucidated on the basis of their spectroscopic data. Compounds 1 and 2 inhibit the in vitro proliferation of the DLD-1 human colorectal adenocarcinoma cell line with IC50 values of 3.7 and 9.8 μM, respectively, and the PANC-1 human pancreatic carcinoma cell line with IC50 values of 6.1 and 13.8 μM, respectively. Neopetrosiquinone A (1) also inhibited the in vitro proliferation of the AsPC-1 human pancreatic carcinoma cell line with an IC50 value of 6.1 μM. The compounds are structurally related to alisiaquinone A, cyclozonarone and xestoquinone.
Keywords: neopetrosiquinone, Petrosia cf. proxima, deep-water, sponge, sesquiterpene quinone, marine natural products, cytotoxic, β-catenin, Tcf-4
Introduction
As part of our program to identify novel marine natural products with potential therapeutic properties, a library of extracts from the Harbor Branch Oceanographic Institute (HBOI) was tested in an assay designed to identify antagonists of β-catenin and T cell factor 4 (Tcf4) binding.1 The interaction between β-catenin and Tcf4 leads to transcription of genes involved in cell proliferation and it has been hypothesized that small molecules that block this interaction may be useful in the treatment of a number of cancers, including colon cancer.2 Screening of the HBOI extract library led to the identification of an organic partition from a deep-water sponge Petrosia cf. proxima that blocked β-catenin/Tcf4 binding (IC50=1.67 μg/mL). This material also showed cytotoxic activity against the PANC-1 pancreatic carcinoma and the DLD-1 colon carcinoma tumor cell lines. Herein we report the isolation, structure elucidation, and biological activity of two novel sesquiterpene benzoquinones, which we have designated as neopetrosiquinones A (1) and B (2) from this sponge.
The sponge material used in this study was collected using the Johnson-Sea-Link I human occupied submersible at a depth of 140 m off the north coast of Jamaica and stored at -20 °C until work up. The frozen sponge was extracted exhaustively with ethanol and after concentration, the residue was partitioned between ethyl acetate and water. The organic partition was further separated by vacuum column chromatography on a silica gel stationary phase using a step gradient of ethyl acetate in heptane to yield eight fractions. The fraction that showed activity in both the β-catenin/Tcf-4 assay and the tumor cell lines was further separated using reverse-phase flash chromatography on an Isco Combiflash™ Companion™ followed by reverse-phase HPLC leading to the isolation of neopetrosiquinone A (1) and neopetrosiquinone B (2).
Results
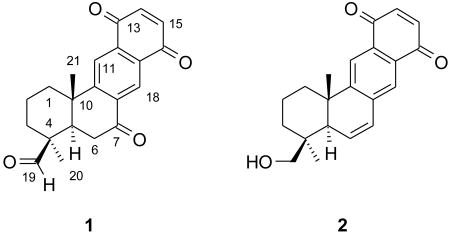
Neopetrosiquinone A (1) was isolated as a yellow oil. Interpretation of the HRDARTMS data coupled with the 13C NMR spectrum suggested a molecular formula of C21H20O4 for 1 [(M+H+) m/z observed 337.1436, calcd 337.1440, Δ = -0.4 mmu], requiring twelve degrees of unsaturation. The 13C NMR spectrum together with the HSQC data suggested the presence of an aldehyde [δH 9.88, δC 204.5, C-19], three ketones [δC 183.9, C-16; 184.5, C-13; 195.0, C-7], eight olefinic carbons of which four are quaternary [δC 130.2, C-12; 134.4, C-8; 134.9, C-17; 159.3, C-9;] and four are methines [δC 123.7, δH 8.16 (s), C-11; δC 126.7, δH 8.64 (s), C-18; δC 139.3, δH 7.0 (AB q, 10.3 Hz), C-14; δC 138.8, δH 7.0 (AB q, 10.3 Hz), C-15]. These resonances accounted for eight degrees of unsaturation and suggested that 1 has four rings. The IR spectrum of 1 displayed adsorption bands at 1726, 1689, and 1664 cm-1 suggesting the presence of aldehyde, conjugated ketone and quinone functionality,3 which was further supported by the UV spectrum, for which maxima were observed at 217, 252, and 337 nm consistent with the presence of a 1,4-napthoquinone moiety.4
The planar structure of neopetrosiquinone A (1) was determined by analysis of the one- and two-dimensional NMR spectra. The 1H-13C one-bond connectivities from the edited-g-HSQC experiment and 1H-1H scalar coupling from the 2D-DQF-COSY spectrum allowed for the assignment of two proton spin systems (See figure S1 in the Supporting Information). The C-1 to C-3 fragment was assigned based on correlations observed in the 2D g-DQF-COSY spectrum as follows: The methylene protons observed at δH 162 (H-1b, m) and 2.55 (H-1a, br dt, 13.1, 3.4Hz) showed coupling to the methylene protons observed at δH 1.82 (H2-2, 2H, m) which in turn were coupled to the methylene protons observed at δH 123 (H-3b, m) and 2.23 (H-3a, dt, 14.4, 3.4 Hz). The C-5 to C-6 fragment was assigned based on a correlation in the DQF-COSY spectrum between the methine proton observed at δH 2.19 (H-5, dd, 14.8, 4.8 Hz) and the methylene protons observed at δH 3.05 (H-6b, dd, 17.9, 14.4 Hz) and 3.14 (H-6a, dd, 17.9, 3.4 Hz).
The HMBC data was instrumental in tying together the remaining structure. Correlations observed in the HMBC spectrum between H2-2, H-3a, and H-6b with the quaternary carbon observed at δC 48.9 (C-4) suggested the connection of the two fragments derived from the COSY spectra through the C-4 quaternary carbon. This was further supported by an HMBC correlation observed between H-3a and the carbon observed at δC 49.2 (C-5). The aldehyde proton observed at δH 9.88 (H-19) showed long range coupling to both C-4 and C-3 in the HMBC spectrum placing it on C-4. A resonance attributable to a methyl singlet observed at δH 1.12 (H3-20) showed long range coupling to C-3, C-4, and C-19 placing it as the fourth substituent on C-4.
In the HMBC spectrum, H-1a, H-1b, and H-6a showed correlations to a quaternary carbon observed at δC 39.0 (C-10). The methyl singlet observed at δH 1.18 (H3-21) showed correlations in the HMBC spectrum to C-1, C-5, and a quaternary carbon observed at δC 159.3 (C-9) consistent with attachment at the quaternary carbon C-10. The correlations observed for H3-21 along with an HMBC correlation observed between H-6a and C-10 provided evidence for the closure of ring A between C-5 and C-10.
Rings B through D were assigned based on correlations observed in the HMBC spectrum. H-6a and H-6b showed correlations to a ketone carbon observed at δC 195.0 (C-7) and to a quaternary olefinic carbon observed at δC 134.4 (C-8). The chemical shifts of H-6a/b are consistent with placement adjacent to the ketone. The proton observed at δH 8.64 (H-18) showed long range coupling to the same ketone carbon [δC 195.0 (C-7)], the quaternary carbon observed at δC 159.3 (C-9), an aromatic carbon observed at δC 134.9 (C-17) and a second ketone observed at δC 183.9 (C-16). A methine proton observed at δH 8.16 (H-11) showed correlations in the HMBC spectrum to the quaternary carbons observed at δC 134.4 (C-8) and 39.0 (C-10) and a weak correlation to the ketone observed at δC 195.0 (C-7). H-11 also showed coupling to a second aromatic carbon observed at δC 130.2 (C-12 or C-17) and a ketone carbon observed at δC 184.5 (C-13). An AB spin system observed at δH 6.99 d, 10.3 Hz, and 6.98 d 10.3 Hz (H-14/H-15) showed correlations in the HMBC spectrum to the carbons observed at δC 130.2 (C-12 or 17), δC 134.9 (C-12 or 17), δC 184.5 (C-13) and δC 183.9 (C-16). These correlations along with the UV data supported the closure of ring D between the two quaternary aromatic carbons C-12 and C-17 to form a 1,4-napthoquinone. All of the HMBC data, chemical shifts and UV data are consistent with the proposed carbon skeleton, 1, which is related to that of cyclozonarone,5 xestoquinone6 and alisiaquinone.7
The relative configuration of 1 was assigned on the basis of interpretation of the 2D-NOESY spectrum and evaluation of the 1H-1H scalar coupling constants. H-5 appears as a dd with J values of 14.8 and 4.8 Hz, which assign it as axial. H-6b appears as a dd with J values of 17.9 and 14.4 Hz, suggesting it is also axial. In the NOESY spectrum, a strong correlation is observed between H-6b and H3-21 which is attributable to a 1,3 diaxial interaction and allowed for the assignment of a trans ring junction. The aldehyde proton H-19 also showed a correlation in the NOESY spectrum with H3-21 and can be attributed to a 1,3 diaxial interaction placing it on the same face of the molecule as H3-21 and H-6b.
Neopetrosiquinone B was isolated as a yellow oil. The HRDARTMS and 13C NMR data suggested a molecular formula of C21H22O3 for 2 [[M+H]+ m/z observed 323.1660, calcd 323.1647, Δ = 1.3 mmu], requiring eleven degrees of unsaturation. The IR spectrum displayed adsorption bands at 1671 cm-1 consistent with the presence of a quinone in 2. Inspection of the NMR spectra of 2 suggested that it is very similar in structure to 1. The major differences included replacement of the C-19 aldehyde functionality with a primary alcohol [δC 66.3 and δH 3.81 (d, J=11.3 Hz) and 3.74 (d, J=11.3 Hz)] and the C-7 ketone functionality with a Δ6-7 double bond [δC7 126.9 and δH7 6.67 (dd J=9.6, 3.4); δC6 134.5 and δH6 6.42 (dd J=9.6, 2.8)]. Resonances attributable to the remaining atoms in the molecule were substantially unchanged and all 2D data (DQF-COSY, HMBC) were consistent with the assignment of the carbon skeleton shown for 2. The relative configuration of 2 was assigned to be the same as for 1 based upon the observation of correlations in the 2D-NOESY spectrum between H2-19 and H3-21 placing them on the same side of the ring, Additional support for the assigned relative configuration was the NOESY correlation observed between the equatorial ring substituents H-6 and H3-20.
Multiple attempts had been made to measure the [α]D for 1 and 2 as well as to obtain a CD spectrum but without success.† Two different polarimeters failed to return a rotation suggesting that the specific rotation is either exceedingly small or that the compounds exist as a racemic mixture. This phenomenon had been observed previously for the sesquiterpene quinone, spongiaquinone8 while neomanuthaquionone has shown a very small specific rotation. Assignment of the absolute configuration of the neopetrosiquinones will require chemical synthesis as no assumptions regarding the absolute configuration can be accurately made based upon the available data.9
Neopetrosiquinones A (1) and B (2) were tested for cytotoxicity in the A549 human lung carcinoma, the PANC-1 and AsPC-1 human pancreatic carcinoma, the DLD-1 human colorectal adenocarcinoma, and the NCI-ADR-Res human ovarian carcinoma cell lines (Table 2). Petrosiquinone A (1) was the more active of the two compounds with moderate cytotoxicity in the DLD-1, PANC-1, and AsPC-1 cell lines. Neopetrosiquinone B (2) displayed marginal cytotoxicity in the DLD-1 and PANC-1 cell lines. Assay data for the pure materials is not available for the β-catenin/Tcf4 assay as the program at Novartis was discontinued prior to final testing of these compounds. In the latter assay, all active fractions contained 1 (with some fractions being > 90% 1) while fractions without this component were not active, suggesting that the aldehyde functionality is important for inhibiting the binding of β-catenin to Tcf4. Related compounds such as halenaquinone did not show activity in this assay.
Table 2.
IC50 values for Neopetrosiquinone A (1) and B (2) against a tumor cell line panel. Values are reported in μM.
| Compound | A549 | PANC-1 | AsPC-1 | DLD-1 | NCI-ADR-Res |
|---|---|---|---|---|---|
| 1 | >14.8 | 6.1 | 6.1 | 3.7 | >14.8 |
| 2 | >15.5 | 13.8 | >15.5 | 9.8 | >15.5 |
Discussion
The neopetrosiquinones represent new members of the sesquiterpene quinone class of secondary metabolite that have been reported from a variety of marine organisms. They are most closely related to cyclozonarone from the brown algae, Dictyopteris undulata, 5xestoquinone from the sponge, Neopetrosia sapra6 and alisiaquinones from an unidentified deep water sponge.7 Cyclozonarone has been reported to have feeding deterrent activity against young abalones and was recently reported to have activity against Trypanosoma cruzi epimastigotes Tulahuen strain (Chagas' disease) with a GI50 value of 700 nM.10 Xestoquinone has shown limited activity in cytotoxicity screens but was found to be a moderate inhibitor of the oncogenic protein tyrosine kinase pp60V-SRC,11 an inhibitor of Ca2+ ATPase from the skeletal muscle myosin,12 and showed moderate in vivo activity in mice infected with Plasmodium berghei NK65.13 Alisiaquionones were reported to have activity against the enzyme targets, plasmodial kinase PFnek-1 and protein farnesyl transferase and against Plasmodium falciparum both in vitro and in vivo.7 In this paper we report that the neopetrosiquinones have moderate cytotoxicity against a panel of tumor cell lines. Biological assay of fractions that were highly enriched in neopetrosiquinone A (up to 90% neopetrosiquinone A) showed activity in an assay designed to detect materials that block the interaction between β-catenin and Tcf4. Only fractions containing neopetrosiquinone A showed activity in this assay and it may be possible that the aldehyde functionality in 1 reacts covalently through imine formation with one of the proteins in the assay thus blocking the protein-protein interaction. Additional work is required to confirm this as well as to determine if neopetrosiquinone blocks the β-catenin/Tcf4 signaling pathway in cell based systems.
Experimental Section
General Experimental Procedures
The IR spectra were collected on a Thermo Nicolet IR100 Spectrometer with potassium bromide discs. The UV spectra were collected on a Hitachi U-3010 spectrophotometer. The optical rotation was attempted on a JASCO DIP-370 digital polarimeter and a Perkin Elmer 343. Reversed-phase C-18 flash chromatography was performed using an Isco Combiflash equipped with PeakTrak software. NMR data were collected on a JEOL ECA-600 spectrometer operating at 600.17 MHz for 1H and 150.9 MHz for 13C. The edited-gHSQC spectrum was optimized for 140 Hz and the gHMBC spectrum was optimized for 8 Hz. Chemical shifts were referenced to solvent, e.g. CD2Cl2, δH observed at 5.32 ppm and δC observed at 54.0 ppm; CDCl3, δH observed at 7.24 ppm and δC observed at 77.2 ppm. The HR-MS was performed using a JEOL AccuTOF DART mass spectrometer.
Biological Material
The sponge used in this study is a specimen of Neopetrosia cf proxima (Phylum Porifera, Class Demospongiae, Order Haplosclerida, Family Petrosiidae; originally described by Zea as Xestospongia proxima;14 and as Neopetrosia by Hooper and Van Soest15). It was collected using Harbor Branch Oceanographic Institute's Johnson-Sea-Link I human occupied submersible (dive number JSL I-3614) off the north coast of Jamaica, St. Ann's Bay (18°27.028'N, 77°10.807'W) at a depth of 140 m on a rock ledge. It was thickly encrusting (∼ 2 cm thick) and had numerous spherical, compound oscules scattered over the surface. The sponge was brownish yellow alive; in ethanol, it is dark brown. The consistency in life was firm; in ethanol, it is easily broken. The ectosome is finely granular and not detachable from the choanosome. The choanosome and base of the sponge incorporate shell fragments. The ectosomal spicules form a dense layer of curved strongyloxeas in one size category, 150-200 μm × 8 μm. The choanosomal spicules are strongyloxeas of the same size as the ectosomal spicules, and less abundant thin oxeas. There are no microscleres. A reference sample preserved in ethanol is deposited in the Harbor Branch Oceanographic Museum (catalog number 003:01037, sample number 7-IX-93-1-008) and is available upon request for taxonomic evaluation.
Extraction and Isolation
The frozen sponge (112 g wet wt) was extracted exhaustively by macerating with ethanol (Pharmco 100%) using a Waring blender (5 × 250 mL). The combined filtered extract was concentrated under reduced pressure to yield 7.59 g of crude residue. The residue was partitioned between ethyl acetate and water (3 × 50 mL portions). The ethyl acetate partition was concentrated under reduced pressure to yield 1.68 g of a dark brown solid. The ethyl acetate partition was fractionated by vacuum column chromatography on a Kieselgel 60 H (EM Science) stationary phase using a step gradient. A 60 mL Büchner funnel fitted with a coarse-porosity fritted glass disk was used as the column. The stationary phase was packed to a total height of 3.5 cm. Fractions were eluted using a 20% step gradient of ethyl acetate in heptane followed by a 100% methanol wash. The eluent series is as follows: fraction 1: heptane (150 mL); fraction 2: heptane-ethyl acetate, 4:1 v/v (150 mL); fraction 3: heptane-ethyl acetate, 3:2 v/v (150 mL); fraction 4: heptane-ethyl acetate, 2:3 v/v (150 mL); fraction 5: heptane-ethyl acetate, 1:4 v/v (150 mL); fraction 6: ethyl acetate (150 mL); fraction 7: methanol (150 mL). Fraction 4 was further separated by reversed phase C-18 medium pressure liquid chromatography on an Isco Combiflash™ Companion™ using a custom packed column [Altec 1.5 cm i.d. × 25 cm column packed with Vydac Protein and Peptide C18 stationary phase (Grace Catalog Number 218TPB 2030 20-30 μ particle size)] and eluted using a nonlinear gradient collected into 13 mm tubes. The elution gradient was: Solvent A: 100% water; Solvent B: 100% CH3CN; t=0 min, A:B (90:10); t=10 min, A:B (80:20) and hold for 20 min; t=30 min, A:B (80:20); t=50 min, A:B (60:40) and hold for 20 min; t=70 min, A:B (60:40); t=80 min, 100% CH3CN and hold for 10 min; flow = 5 mL/min; detected by UV at 240 nm. Fraction 9 (tube 42) was further purified by reverse-phase HPLC using a Vydac Protein and Peptide C18 column (10mm × 250 mm, 10 μm particle size), flow rate 3 mL/min; eluent: H2O-CH3CN, 57:43; monitored by UV at 230 nm to give neopetrosiquinone A (1) (0.0034 g, 3.0 × 10-3 % of frozen weight) and neopetrosiquinone B (2) (0.0016 g, 1.4 × 10-3 % of frozen weight).
Neopetrosiquinone A (1)
yellow oil; [α]20D unable to measure (c 0.14 MeOH:CH2Cl2, 9:1); λmax (log ε) 217 (4.45), 252 (4.30), 337 (3.44) nm; IR (KBr) vmax 2886, 2701, 1726, 1689, 1664, 1595, 1259, 1149, 1053, 847 cm-1; 1H and 13C NMR (Table 1); HRMS: (M + H+) m/z 337.1436 observed, calcd for C21H21O4, 337.1440.
Table 1. 1H and 13C NMR Data for 1 (CD2Cl2) and 2 (CDCl3), 600 MHz.
| Neopetrosiquinone A (1) | Neopetrosiquinone B (2) | ||||||
|---|---|---|---|---|---|---|---|
| no. | δC, mult. | δH, (J in Hz) | HMBCa | δC, mult. | δH, (J in Hz) | HMBCa | |
| 1 | A | 37.3, CH2 | 2.55, br dt (13.1, 3.4) | 2, 5, 10 | 36.1, CH2 | 2.33, br m | 2, 3, 5, 21 |
| B | 1.62, m | 2, 10 | 1.75, br m | ||||
| 2 | 18.8, CH2 | 1.82, m | 4 | 18.7, CH2 | 1.75, br m | 4, 10 | |
| 3 | A | 34.3, CH2 | 2.23, dt (14.4, 3.4 | 1, 5, 19 | 35.3, CH2 | 1.88, br d (15.1) | |
| B | 1.23, m | 2, 4, 19, 20 | 1.11, br m | 2 | |||
| 4 | 48.9, qC | 38.6, qC | |||||
| 5 | 49.2, CH | 2.19, dd (14.8, 4.8) | 3, 4, 6, 19, 20 | 50.6, CH | 2.29, t (3.4) | 4, 6, 19, 20, 21 | |
| 6 | Ab | 35.4, CH2 | 3.14, dd (17.9, 3.4) | 4, 7, 8, 10 | 134.5, CH | 6.42, dd (9.6, 2.8) | 4, 5, 8 |
| Bc | 3.05, dd (17.9, 14.4) | ||||||
| 7 | 195.0, qC | 126.9, CH | 6.67 dd (9.6, 3.4) | 5, 9 | |||
| 8 | 134.4, qC | 138.7, qC | |||||
| 9 | 159.3, qC | 153.8, qC | |||||
| 10 | 39.0, qC | 38.5, qC | |||||
| 11 | 123.7, CH | 8.16, s | 7,8,10, 12, 13 | 120.8, CH | 7.87, s | 8, 10, 13/16, 17 | |
| 12 | 134.9, qCd | 131.2, qCd | |||||
| 13 | 184.5, qC | 185.2, qC | |||||
| 14 | 138.8, CHe | 6.99, d (10.3)e | 12, 13, 16, 17 | 139.1, CHe | 6.91, d (10.3) e | 13/16 | |
| 15 | 139.3, CHe | 6.98, d (10.3) e | 12, 13, 16, 17 | 138.6, CHe | 6.89, d (10.3) e | 13/16 | |
| 16 | 183.9,qC | 185.2, qC | |||||
| 17 | 130.2, qCd | 130.5, qCd | |||||
| 18 | 126.7, CH | 8.64, s | 7, 9, 16, 17 | 124.3, CH | 7.70, s | 7, 9, 12, 13/16 | |
| 19 | A | 204.5, CH | 9.88, s | 3, 4, 5 | 66.3, CH2 | 3.81, d (11.7) | 20 |
| B | 3.74, d (11.0) | 3, 20 | |||||
| 20 | 22.5, CH3 | 1.12, s | 3, 4, 19 | 26.3, CH3 | 1.09, s | 3, 4, 5, 19 | |
| 21 | 23.5, CH3 | 1.18, s | 1, 5, 9 | 20.8, CH3 | 1.06, s | 1, 5, 9, 10 | |
HMBC correlations, optimized for 8 Hz, are from proton(s) stated to the listed carbon(s);
Equatorial proton;
Axial proton;
Assignments may be interchanged;
AB spin system, assignments may be interchanged
Neopetrosiquinone B (2)
yellow oil; [α]20D unable to measure (c 0.13 MeOH:CH2Cl2, 9:1); λmax (log ε) 216 (4.35), 255 (4.13), 365 (3.27) nm; IR (KBr) vmax 3464, 2935, 2880, 1671, 1595, 1314, 1053, 853, 737 cm-1; 1H and 13C NMR (Table 1); HRMS: (M + H+) m/z 323.1660 observed, calcd for C21H23O3, 323.1647.
Biological Assays
Neopetrosiquinone A and B were evaluated for their effects on the proliferation of A549 human lung adenocarcinoma (ATCC No. CCL-185), PANC-1 human pancreatic cancer (ATCC No. CRL-1469), AsPC-1 human pancreatic cancer (ATCC No. CRL-1682), DLD-1 human colorectal adenocarcinoma (ATCC No. CCL-221), and NCI-ADR-RES (formerly MCF-7/ADR) human ovarian carcinoma. The A549, PANC-1, AsPC-1, and DLD-1 cell lines were obtained from the American Type Culture Collection (Rockville, MD). The NCI-ADR-RES cell line was obtained from the NCI-Frederick Cancer DCTD Tumor/Cell Line Repository (Bethesda, MD). Cytotoxicity assays were run using protocols described previously.16 The β-catenin/TCF4 binding assay used in this study have been described previously.1 In brief the assay was run as follows: Purified β-catenin (amino acids 134–668) was coated on microtiter plates and incubated sequentially with a Tcf4 fragment (residues 8–54) fused to glutathione-S-transferase (GST), anti-GST antibody, and alkaline phosphatase (AP) conjugated secondary antibody. Compounds that disrupt the Tcf/β-catenin complex thus register reduced AP values relative to the background.
Supplementary Material
Acknowledgments
Partial funding for the research described in this manuscript was provided by grants from the NIH including 2U19-CA529955 and 2RO1-CA093455. A graduate fellowship to PW was provided by the State of Florida Center of Excellence for Biomedical and Marine Biotechnology. We thank A. Wood of Novartis Institutes for Biomedical Research for the β-catenin/Tcf4 assay results. We thank C. Cody of JEOL for providing HRMS data. This is HBOI Contribution Number XXXXXX.
Footnotes
Neopetrosiquinones A and B are highly colored. Initially, it was considered that the color of the sample may not have allowed light to pass through the cell but upon numerous dilutions, no stable reading could be reached.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lepourcelet M, Chen YN, France DS, Wang H, Crews P, Petersen F, Bruseo C, Wood AW, Shivdasani RA. Cancer Cell. 2004;5:91. doi: 10.1016/s1535-6108(03)00334-9. [DOI] [PubMed] [Google Scholar]
- 2.van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, Tjon-Pon-Fong M, Moerer P, van den Born M, Soete G, Pals S, Eilers M, Medema R, Clevers H. Cell. 2002;111:241. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 3.Pretsch E, Clerc T, Seibl J, Simon W. Tables of Spectral Data for Structure Determination of Organic Compounds: 13C-NMR, 1H-NMR, IR, MS, UV/VIS Chemical Laboratory Practice. Springer-Verlag; 1983. [Google Scholar]
- 4.Rodrigues S, Viana L, Baumann W. Analytical and Bioanalytical Chemistry. 2006;385:895. doi: 10.1007/s00216-006-0502-6. [DOI] [PubMed] [Google Scholar]
- 5.Kurata K, Taniguchi K, Suzuki M. Phytochemistry. 1996;41:749. [Google Scholar]
- 6.Nakamura H, Kobayashi Ji, Kobayashi M, Ohizumi Y, Hirata Y. Chem Lett. 1985:713. [Google Scholar]
- 7.Desoubzdanne D, Marcourt L, Raux R, Chevalley S, Dorin D, Doerig C, Valentin A, Ausseil F, Debitus C. J Nat Prod. 2008;71:1189. doi: 10.1021/np8000909. [DOI] [PubMed] [Google Scholar]
- 8.Capon RJ, Groves DR, Urban S, Watson RG. Aust J Chem. 1993;46:1245. [Google Scholar]
- 9.Yong KWL, Jankam A, Hooper JNA, Suksamrarn A, Garson MJ. Tetrahedron. 2008;64:6341. [Google Scholar]
- 10.Cuellar MA, Salas C, Cortés MJ, Morello A, Diego Maya J, Preite MD. Bioorganic & Medicinal Chemistry. 2003;11:2489. doi: 10.1016/s0968-0896(03)00193-7. [DOI] [PubMed] [Google Scholar]
- 11.Lee RH, Slate DL, Moretti R, Alvi KA, Crews P. Biochem Biophys Res Commun. 1992;184:765. doi: 10.1016/0006-291x(92)90656-6. [DOI] [PubMed] [Google Scholar]
- 12.Sakamoto H, Furukawa KI, Matsunaga K, Nakamura H, Ohizumi Y. Biochemistry. 1995;34:12570. doi: 10.1021/bi00039a011. [DOI] [PubMed] [Google Scholar]
- 13.Laurent D, Jullian V, Parenty A, Knibiehler M, Dorin D, Schmitt S, Lozach O, Lebouvier N, Frostin M, Alby F, Maurel S, Doerig C, Meijer L, Sauvain M. Bioorg Med Chem. 2006;14:4477. doi: 10.1016/j.bmc.2006.02.026. [DOI] [PubMed] [Google Scholar]
- 14.Zea S. Esponjas del Caribe Colombiano, Catalogo Cientifico. Santa Marta, Colombia: 1987. [Google Scholar]
- 15.Hooper JNA, VS RWM, editors. Systema Porifera, A guide to the classification of sponges. Kluwer Academic; New York: 2002. [Google Scholar]
- 16.Gunasekera SP, Zuleta IA, Longley RE, Wright AE, Pomponi SA. J Nat Prod. 2003;66:1615. doi: 10.1021/np030292s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


