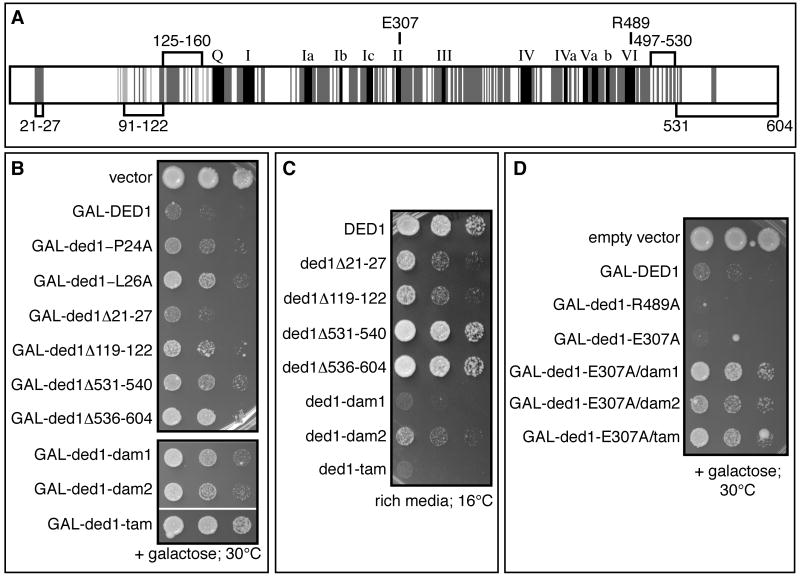
Figure 1. Genetic approach to identify separation-of-function alleles of DED1.
A. The conserved motifs of the DEAD-box protein family are shown in black (Fairman-Williams et al., 2010). Conservation among Ded1 orthologs is shown in dark grey (from yeast to humans) or light grey (among fungi). The numbers mark amino acid domains of Ded1 that are implicated in promoting translation (amino acids labeled above schematic) or in promoting assembly of a Ded1-eIF4F-mRNA complex (amino acids labeled below the schematic). B. Effects of over-expression of ded1 alleles from a galactose inducible promoter in the presence of wild-type, endogenous Ded1. C. Growth of yeast expressing single copies of wild type or mutant ded1 at low temperature. Strains containing a plasmid with wild type or mutant DED1 under the control of its endogenous promoter as the sole copy of DED1 (yRP2799) were spotted at the same concentration on rich media and grown at 16°C for 7 days. D. Cells were treated as in B. See also Table S4 and Figure S2.