Abstract
Background and Aims
Grevillea rhizomatosa is a spreading shrub which exhibits multiple breeding strategies within a narrow area in the fire-prone heathlands of eastern Australia. Reproductive strategies include self-compatibility, self-incompatibility and clonality (with and without sterility). The close proximity of contrasting breeding systems provides an opportunity to explore the evolution of sterility and to compare and contrast the origins of genotypic diversity (recombinant or somatic) against degrees of sexual expression.
Methods
ISSR markers for 120 band positions (putative loci) were used to compare genetic diversity among five populations at a macro-scale of 5 m between samples (n = 244 shrubs), and at a micro-scale of nearest neighbours for all plants in five 25-m2 quadrats with contrasting fertilities (n = 162 shrubs). Nearest-neighbour sampling included several clusters of connected ramets. Matrix incompatibility (MIC) analyses were used to evaluate the relative contribution of recombination and somatic mutation to genotype diversity.
Key Results
High levels of genotypic diversity were found in all populations regardless of fertilities (fertile populations, G/N ≥ 0·94; sterile populations, G/N ≥ 0·97) and most sterile populations had a unique genetic profile. Somatic mutations were detected along connected ramets in ten out of 42 ramet clusters. MIC analyses showed that somatic mutations have contributed to diversity in all populations and particularly so in sterile populations.
Conclusions
Somatic mutations contribute significantly to gene diversity in sterile populations of Grevillea rhizomatosa, the accumulation of which is the likely cause of male and female sterility. High levels of genetic diversity therefore may not always be synonymous with sexual fitness and genetic health. We hypothesize that frequent fires drive selection for clonal reproduction, at the cost of flowering such that sexual functions are not maintained through selection, and the build-up of somatic mutations in meristems results in high genotype diversity at the cost of pollen and ovule fertilities.
Keywords: Clone diversity, ISSR, matrix incompatibility, multiple breeding strategies, sterility, Grevillea rhizomatosa, somatic mutations
INTRODUCTION
The world is replete with examples of rare and endangered plant species on the brink of extinction (wild macadamias, for example; Pisanu et al., 2009; Neal et al., 2010); however, in the face of human-induced adversity there are also many rare plant species that persist in the landscape (e.g. cliff-dwelling Bertya ingramii; Scott and Gross, 2004). Long-livedness is one life-history trait that may promote the persistence of populations through individuals living long and having many opportunities to reproduce thus compensating for times where seed production or establishment are not favourable. In support of this, Sjöström and Gross (2006), suggest that a lack of trees on the recently extinct plant database for Australia is a direct result of the long-livedness of the tree habit imparting resilience to populations of such species. Indeterminate growth through clonal reproduction may also afford longevity and there are several remarkable examples of seemingly ‘immortal’ individuals (see table 1 in de Witte and Stöcklin, 2010). However, the costs of indeterminate growth, where meiosis is avoided and, as the clone spreads, the number of mitotic cell divisions increases, may include an accumulation of mutations. The accumulation of mutations is likely to buoy the level of genetic diversity in clonal individuals as deleterious mutations are less likely to be purged in the absence of sexual recombination. Indeed there are many examples where clonal organisms have high levels of gene diversity (Ellstrand and Roose, 1987), although the benefits, if any, of this are rarely tested and require assessments of asexual fitness (e.g. Ally et al., 2010).
Since Muller's work (Muller's Ratchet; Felsenstein, 1974) in 1964 and Klekowski's somatic mutation theory of clonality in 1997 (Muller, 1964; Klekowski, 1997), there has been a growing interest in the study of sterility in clonal plants (e.g. Eckert, 2002). One model pathway for the advent of sterility is that if the sexual machinery is not under active selection (i.e. due to a lack of fertilization) and vegetative reproduction is possible and dominates, background somatic mutations are not purged but build up to have the effect of disrupting function including processes critical to sexual fertility (e.g. Eckert et al., 1999). The insertion of T-DNA through mutagenesis in the MAP3Kɛ1 and MAP3Kɛ2 protein kinase genes of Arabidopsis, for example, has been shown to cause plasma membrane irregularities following pollen mitosis I which causes pollen lethality (Chaiwongsar et al., 2006).
Less work has been undertaken on the environmental mechanisms that may promote clonal growth. In landscapes where frequent wildfire prevents individuals from flowering and fruiting, and clonal growth is promoted, the scene is set for somatic mutations to build up in the absence of purging. Such mutations may disable sexual recombination such that when flowering is permitted during fire-free periods, seed cannot be produced. The types of species prone to this could include those that inhabit fire-prone ecosystems, which take several years to reach sexual maturity, require several years of a fire-free period to re-initiate flowering and resprout after fires. Many species in the Proteaceae exhibit these life history traits and exist in fire-prone habitats. The growing membership of proteads with infertile, obligate-clonal species in Grevillea (Kimpton et al., 2002; Holmes et al., 2008, 2009), Lomatia (Lynch et al., 1998), Banksia (Millar et al., 2010) and Hakea (Smith, 2004), for example, is thus of great interest. Where entire populations are infertile and obligately clonal, somatic mutations may accrue, resulting in a very different genetic profile than sexually reproducing populations of the same species. However, studies incorporating both sterile and fertile populations are few for woody species with bisexual flowers, and lacking in the large southern hemisphere family, Proteaceae. We set out to compare genetic profiles across different sexual strategies using the threatened species Grevillea rhizomatosa, a long-lived shrub that has self-incompatible, self-compatible and fully sterile populations, all of which occur in a fire-prone habitat and have individuals that reproduce vegetatively (Gross and Caddy, 2006). Our first objective was to evaluate and compare levels of divergence in gene diversity in sterile populations compared with that found in nearby sexually reproducing populations. We expected overall genotypic diversity to be highest in self-incompatible populations, followed by the self-compatible population and lowest in sterile populations. Information on levels of divergence can inform estimations of clone age (Ally et al., 2008) and speciation trajectories (Vanormelingen et al., 2008) and, when multiple breeding systems are present, it can provide information on life-history strategies and population dynamics.
Our second objective was to determine if somatic mutations are frequent events in Grevillea rhizomatosa. The unequivocal detection of somatic mutations for wild species can be an elusive task. It is dependent upon a variable molecular marker and a suitable target organism (ideally one with rapid vegetative reproduction) with ramets that can be distinguished (e.g. with extant connections between ramets). This is the problem of ‘being at the right place at the right time with the right tools’. As it can be difficult to satisfy all of these requirements, the likelihood of making incorrect conclusions (i.e. incorrectly excluding somatic mutagenesis as a contributor to gene diversity) is an issue. Several approaches were employed to try and minimize the risk of these Type I errors including the excavation of ramets. We aimed to determine if somatic mutations are more common in sterile compared with fertile populations where purging through recombination and natural selection would be more frequent.
MATERIALS AND METHODS
Species and study sites
Grevillea rhizomatosa Olde & Marriott (Proteaceae) is a spreading shrub growing to 1·5 m in height that flowers sporadically throughout the year but mainly in August–November. It is restricted to a 27-km2 area on the Gibraltar plateau (see fig. 1 in Caddy and Gross, 2006) where it often grows adjacent to creeks on granitic soil. Reproduction occurs through the proliferation of rhizomatous suckers and less often by seed production (Gross and Caddy, 2006). Ramets are produced from subterranean stems or thickened roots that can extend below ground for several metres before a stem sucker emerges above ground. Often lines of smaller plants can be seen emerging from a central plant (a guerilla strategy). Excavations revealed that these are stem ramets and not root fusions between genets. The suckering habit of G. rhizomatosa enables rapid regeneration after fire (i.e. resprouter). As such, populations are comprised of individuals that are of an indeterminate age and plants in small patches could be derived from the same genet.
Six populations within a 9 km × 3 km area were studied within the contiguous national parks of Gibraltar Range and Washpool in north-eastern NSW, Australia (29°28′03″S to 29°32'37″S, 152°18'18″E to 152°21′39″E; see fig.1 in Caddy and Gross, 2006). Populations are small, each with fewer than 300 plants: Washpool (approx. 225 individuals), Dandahra Trail (approx. 250 individuals), Cascade (41 individuals), Swamp (165 individuals), Mulligan's Hut (approx. 250 individuals) and Cascade Walk (approx. 80 plants). The Mulligan's Hut and Swamp populations are sterile (Caddy and Gross, 2006) as are the plants in the Cascade Walk population (C. L. Gross, unpubl. res.). The Cascade and Dandahra Trail populations have a self-incompatibility mechanism that is absent from the Washpool population (Gross and Caddy, 2006). We do not know if the incompatibility mechanism is the result of an S-allele system or the result of early-acting inbreeding depression. In the Washpool population seed set can occur from assisted self-pollination, autogamous pollination as well as from outcrossed pollen (Gross and Caddy, 2006). The species is diploid in both sterile and fertile populations (2n = 20; Nelson, 2006).
Genetic diversity at macro- and micro-scales
Genotypic diversity was investigated at two scales. At a macro-scale we aimed to quantify the number of genotypes in the greater study area by sampling extensively from five populations. Using a micro-scale survey we looked for genetic variation in small neighbourhoods (NH = all plants within a 25-m2 quadrat) where groups of plants were likely to contain ramets. Here our aim was to detect the extent of clonality in small patches and whether connected plants were of the same genotype. Field sampling was undertaken between 2006 and 2010.
Macro-scale genetic variation
Leaf material was collected from at least 35 plants in each of five populations (Washpool, n = 54 plants; Dandahra trail, n = 50 plants; Cascade, n = 50 plants; Swamp, n = 35 plants; Mulligan's Hut, n = 55 plants). To minimize sampling the same genet, leaf tips were taken from individuals that were separated by at least 5 m. Samples were labelled and dried in silica gel in preparation for DNA extraction.
Micro-scale genetic variation
Neighbourhood quadrats (NH = all plants in a 25-m2 quadrat were sampled) were set out in three populations. Populations that had a high risk of erosion from excavation work were not used. Within quadrats, nearest neighbour distances were measured so that neighbourhood maps could be constructed. Sampled plants were checked for connections to other plants through careful excavations. Two neighbourhoods were sampled in fertile and self-incompatible Cascade (Cas–NH1 = 25 plants, Cas–NH2 = 23 plants), one in sterile Cascade Walk (CasW–NH3 = 56 plants) and two in sterile Mulligan's Hut (Mull–NH4 = 32 plants, Mull–NH5 = 26 plants). At Cascade the two quadrats were separated by a creek. At Mulligan's Hut the quadrats were approx. 30 m apart and separated by boulders. Leaf tips (10 cm) were taken from every shrub in the quadrat (25 m2). Samples were labelled and dried in silica gel in preparation for DNA extraction.
DNA extraction
Apical shoots (20 mg) were placed in 2-mL centrifuge tubes with a 3-mm tungsten carbide bead and pulverized in a mixer mill for 30 min, followed by a digestion with 400 µL AP1 buffer (DNeasy plant mini kit; Qiagen) for 30 min at 65 °C. Then 130 µL AP2 buffer was added and the mixture incubated on ice for 5 min and centrifuged at high speed for 8 min. From each sample, 200 µL was placed in a reaction plate and total genomic DNA was extracted by the X-tractor Gene robotic system (Corbett) using a standard protocol (Pusterla et al., 2008). The quality of DNA in samples was tested on a 1% agarose gel with standard concentrations of Lambda DNA (1, 20, 50 and 100 ngμL−1) followed by visual assessment of each sample. DNA was quantified using a spectrophotometer (Nanodrop ND-1000; Nanodrop Technologies) and diluted to a concentration of 20 ngμL−1 in PCR-grade water. Twenty-six ISSR primers (Godwin et al., 1997; Wolfe et al., 1998) based on the UBC Primer set 9 (obtained from GeneWorks, Melbourne, Australia) were tested on four individuals from each of six populations. Four primers that generated polymorphic and reproducible bands were selected. Primer 809 [(AG)8G] yielded 30 bands, primer 840 [(GA)8YT] yielded 38 bands, primer 841 [(GA)8YC] yielded 31 bands and primer 888 [BDB(CA)7] yielded 21 bands. These primers were then used for amplification of all samples taken across all populations. The reaction mixture comprised 10 µL of GoTaq® Green Master Mix (Promega), 1 µL of primer (10 µm), 1 µL of template DNA (20 ng) and 8·0 µL of PCR-grade water for each sample to give a total volume of 20 µL. The PCR programme consisted of an initial denaturation at 94 °C for 10 min and each cycle then comprised a 45-s denaturation phase at 94 °C, a 45-s annealing phase at 41 °C, and a 2-min extension phase at 72 °C. After 40 cycles there was a final extension phase of 10 min at 72 °C. The PCR products were then analysed using a multicapillary electrophoresis system with a modified 0M500 method file (QIAxcel DNA High Resolution Kit). To check for DNA extraction repeatability and to set the thresholds of similarity, two samples were extracted separately from the same leaf for six leaves, one leaf from each population. ISSR band outputs were then scored automatically using the QIAxcel Bio Calculator (with thresholds for similarity set at baseline filter = 100 rfu, threshold = 15%, minimum distance = 2·00 bp). Then each sample profile was checked visually for miscalled or poorly identified peaks which were eliminated. The QIAxcel Bio Calculator was used to produce a presence/absence binary score for each sample. Due to a large number of samples, a second stage of data quality-control was undertaken by searching the data matrix for miscalled data. This was done by checking the data for (a) any two bins separated by <5 bp; (b) paired bins (size brackets), which were then merged if they were separated with <5 bp or separated if differences were >5bp; (c) bins with <20 fragments; and (d) all bins which were fully populated across a population were double checked against their respective run files to verify them. Miscalled data occurred in <2 % of the data matrix. In addition the alignments between samples from different analyses were calibrated by shuffling samples and repeating the analyses. This process was repeated for each primer across the entire dataset. In addition and where somatic mutations were detected, samples were re-run and visualized on agarose gels using the methods in Fatemi and Gross (2009).
Statistical analysis
Using the protocols of Fatemi and Gross (2009), bands were scored as either present (1) or absent (0) for all individuals. Analyses were conducted using the binary option for diploid organisms in GenAlEx 6·1 (Peakall and Smouse, 2006). The number of private bands (bands unique to a single population), percentage polymorphism and Shannon's Information Index were calculated at both the macro- and micro-population levels. The distribution of genetic variability was assessed by conducting an analysis of molecular variance (AMOVA). The relationship between individuals and populations was investigated by principal co-ordinate analysis (PCoA).
Clonal or genotypic diversity was estimated by calculating the following two parameters in the macro- and micro-scale populations: (1) the proportion of distinguishable genotypes was measured as G/N, where G is the number of genotypes detected and N is the total number of individuals (ramets) sampled; (2) Simpson's Diversity Index (D) corrected for finite sample sizes by Pielou (1969) was calculated as:
where ni is the number of ramets of ith genotype and N is the sample size. The index D is 0 in a population with only one genet and 1 in a population comprised of genets with only one member. The evenness measure of Fager (1972) was calculated as:
where Dmin = [(G – 1)(2N – G)]/[N(N – 1)] and Dmax = [N(G – 1)]/[G(N – 1)]. E is 0 in a population where all samples represent different genets or where all samples belong to the same genet. E = 1 when there is an even distribution of genotypic diversity (i.e. where all genets are replicated an equal number of times in a population). To estimate the number of genotypes in populations, similarity thresholds were also used for both the macro- and micro-sampling scales by calculating average genetic distances among ramets to provide percentage identity. Scoring errors and mutations may cause individuals from the same clonal lineage (clone-mates) to have a pairwise distance larger than zero. Using the program GenoDive Version 2·0b20 (software for analysis of population genetic data, 2011; see Meirmans and Van Tienderen, 2004) ramets were assigned to genotypes at the 100 % identical (threshold 0) and thresholds 1, 2, 3 and 4 which are based on genetic distances calculated from band profiles. The most conserved limit for allocating clones is threshold 0 (i.e. to be considered a clone-mate, finger-print profiles have to be 100 % identical between pairs) and commonly used limits range from 2 % to 4 % (i.e. any pair of individuals having <2–4 % differences in the profiles are classified as clone-mates; see also Lo et al., 2010, and references therein).
Finally, matrix incompatibility analyses (Mes, 1998) was used to evaluate if genotype profiles were generated via somatic mutations or from recombination events. With binary ISSR data the presence or absence of bands at two loci presents four possible combinations (0/0, 1/0, 0/1, 1/1). The presence of all four combinations is more parsimoniously explained by sexual recombination than by three mutation events, assuming the infinite allele model. The level of incongruence among profiles is referred to as ‘incompatibility’. The incompatibility value is used as a measure of recombination when summed over all pairwise comparisons. The jackknifing JACTAX option in PICA (Wilkinson, 2001) was used to determine the contribution of a particular genotype to matrix incompatibility. The genotypes with the highest contribution were sequentially removed from the dataset until the matrix incompatibility count (MIC) became zero or only four multilocus genotypes were left in the analyses. The number of steps (genotypes) required to reduce the MIC to zero is an indication of the origins of the genetic diversity. If the deletion of most genotypes is required to remove matrix incompatibility, indicative of recent recombination events, there should be a gradual decrease in MIC until only four multilocus genotypes are left in the analyses. If only a few genotypes contribute to overall matrix incompatibility, there should be a sharp decrease in MIC upon deletion of these genotypes, indicating that the majority of genotypes were derived from a pathway involving somatic mutation. These analyses were conducted at a population level for all macro- and micro-scales. In the micro-scale analysis the Cascade Walk neighbourhood (CasW–NH3, n = 56 plants) the quadrat was split into two, giving two batches of 28 samples so that the MIC analysis with the other neighbourhoods was comparable in terms of sample size.
RESULTS
Macro-scale genetic variation
At the macro-scale, 244 shrubs over five populations were sampled. ISSR markers provided 120 band positions of which all were polymorphic across populations, although within populations per cent polymorphisms ranged from 48 % to 65 % (Table 1). At the conservative level of 100 % similarity (where identical genotypes have to share exact matches at each band position) there were 243 genotypes present in the 244 sampled individuals (Table 1). No genotypes were shared among populations and private bands were detected in all populations (Table 1). Genotypic diversity (G/N) was high within populations (0·97–1·00) regardless of fertility levels, and Simpson's Diversity Index showed that all genets were made up of one member except at Swamp where 90 % of the population was made up of one member (Table 1). An evenness value of 0 was found in all macro-populations (Swamp was 0·000000330), indicating that the plants represent different genets. In fact, clones (plants with identical genotypes) were only detected in Swamp (Table 1). The self-incompatible populations (Cascade and Dandahra Trail) exhibited levels of polymorphisms and Shannon's Information Indices comparable to the range found within the sterile population at Swamp. The sterile Mulligan's Hut plants had the lowest detected levels of polymorphic band positions and a low Shannon's Information Index (Table 1).
Table 1.
Summary of genetic/genotypic data collected in populations of G. rhizomatosa at the macro-scale of sampling with at least 5 m between plants within populations
| No. of genotypes (no. of plants in each clone network) at threshold levels 0–4 |
At threshold 0 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Population | n | 0 | 1 | 2 | 3 | 4 | Private bands | P% | G/N | I | D | E |
| Cas–SI | 50 | 50 (1) | 49 (1–2) | 47 (1–2) | 46 (1–3) | 39 (1–4) | 3 | 50 | 1·00 | 0·19 | 1·0 | 0·0 |
| Dand–SI | 50 | 50 (1) | 50 (1) | 50 (1) | 49 (1–2) | 49 (1) | 6 | 65 | 1·00 | 0·24 | 1·0 | 0·0 |
| Mull–sterile | 55 | 55 (1) | 53 (1–2) | 48 (1–3) | 40 (1–11) | 29 (1–15) | 3 | 48 | 1·00 | 0·16 | 1·0 | 0·0 |
| Wash–SC | 54 | 54 (1) | 54 (1) | 54 (1) | 53 (1–2) | 53 (1–2) | 2 | 62 | 1·00 | 0·21 | 1·0 | 0·0 |
| Swamp–sterile | 35 | 34 (1–2) | 32 (1–4) | 32 (1–4) | 31 (1–4) | 29 (1–4) | 1 | 57 | 0·97 | 0·21 | 0·9 | 0·0 |
| Total | 244 | 243 | 238 | 231 | 219 | 199 | 15 | |||||
n = number of plants sampled.
No. of genotypes = number of different genotypes [based on thresholds of 0 (100 % matching bands) and thresholds 1–4 which indicate the maximum genetic distance that is allowed between two individuals to still be clone-mates with the same multilocus genotype]. The size of the clone network is given in parenthesis as the number of clone-mate plants.
Private bands = the number of bands unique to the population; P% = percentage of bands that were variable within populations; G/N = proportion of distinguishable genotypes; I = Shannon's Information Index; D = Simpson's Diversity Index; E = Genotypic evenness measure of Fager.
Populations: Cascade (Cas), Dandahra (Dand), Mulligan's Hut (Mull), Washpool (Wash) and Swamp.
SI = plants are self-incompatible; SC = plants are self-compatible; sterile = flowers male and female sterile.
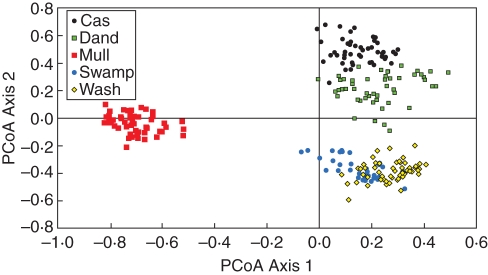
The overall area occupied by a population on the PCoA reflects the amount of genetic variability contained within that population. A large area of occupancy on the figure indicates high variability and overlapping individuals indicate shared genotypic profiles. The population groupings from the PCoA (Fig. 1) showed that populations form aggregated entities with some overlap. Plants from the Mulligan's Hut population formed a discrete group. The self-incompatible populations were grouped together loosely and had a pairwise Nei's Genetic Identity of 0·928. The self-compatible plants from Washpool overlapped in gene diversity with sterile Swamp plants (Fig. 1) and had a pairwise Nei's Genetic Identity of 0·945. The PCoA showed that the Mulligan's Hut population is the most dissimilar to the other populations (Fig. 1) and it had the lowest pairwise Nei's Genetic Identities with the other populations (range 0·884–0·889). The clumped array of genotypes from the Swamp and Washpool populations indicates that there is a comparatively high level of similarity among individuals from those populations (Fig. 1). AMOVA indicated that the majority of genetic variability (54 %) was located within populations (ΦPT = 0·458, P = 0·010).
Fig. 1.
Principal co-ordinate analyses of ISSR data collected from individuals sampled from five populations of G. rhizomatosa in the Washpool and Gibraltar Range National Parks, NSW (PC1 = 31·62 % variation, PC2 = 24·18 % variation, PC3 = 16·68 % variation). Populations are Cascade (Cas), Dandahra (Dand), Mulligan's Hut (Mull), Swamp and Washpool (Wash).
Micro-scale genetic variation
At the micro-scale, 162 shrubs over three populations and five neighbourhoods were sampled and 95 band positions resolved. We detected 146 genotypes and none was shared among neighbourhoods. All neighbourhoods had private bands with sterile Cascade Walk containing the highest number of private bands (Table 2). Genotypic diversity (G/N) in the micro-scale population was high and ranged from 0·77 to 1·00 in sterile neighbourhoods (Table 2) and was 1·00 in both fertile neighbourhoods at Cascade (Table 2). Simpson's Diversity D and Fager's Eveness E where 1·0 and 0, respectively, in the fertile neighbourhoods, indicating that every plant was a unique genotype. In the sterile neighbourhoods, less diversity and less eveness in genotypes was found in D and E, indicating the presence of clones (Table 2). In Mulligan's Hut NH4, plant pairs 2a and 2b and 31a and 31b were each joined below ground by a root-bearing stem (joined ramets) and within pairs the same genotype was found. No plants were joined in the Mulligan's Hut NH5 quadrat and this neighbourhood had the lowest levels of polymorphic bands (Table 2). In the Cascade Walk neighbourhood, where 56 shrubs were sampled, 43 genotypes were detected (Table 2) with five individuals sharing one genotype. In this neighbourhood our sampling included 36 plants that were connected below ground to one or more plants [15 plant clusters of 2 plants (n = 11) or 3 plants (n = 3) or 5 plants (n = 1)]. Six of the 15 ramet clusters had at least one band difference (Figs 2 and 3) while the remaining nine clusters did not express different genotypes on connected pairs of plants. At the Cascade site the two neighbourhoods were separated by a creek. Cascade NH1 and Cascade NH2 had similar levels of gene diversity (Table 2) and were weakly differentiated (Fig. 4), although there were no shared genotypes between these two neighbourhoods. On two occasions in Cascade NH1 we found joined ramets, each involving two plants joined below ground. In one pair, the ramets expressed different genotypes at one band position and in the second joined pair two genotypes were detected, differing at two band positions. At Cascade NH2 we found two pairs of plants that were connected below ground. One pair expressed a different genotype by one band difference and the other pair was of the same genotype. Across all neighbourhoods, ten pairs of connected plants were found where each pair member held a different genotype. Threshold analyses (Tables 1 and 2) provided scenarios of decreased genotypic diversity when the threshold limits from 0 were relaxed to thresholds 1–4. These models suggest that we may have overestimated the number of genotypes in our populations at threshold 0 if, for example, the marker was prone to reading errors, and consequently we may have overestimated the incidence of connected ramets having different genotypes. However, many of the ten plant pairs that expressed different genotypes were still distinguished as separate genotypes at the higher threshold limits of threshold 1 (7/10 pairs), threshold 2 (5/10 pairs), threshold 3 (4/10 pairs) and threshold 4 (2/10 pairs). Furthermore, repeat assays of suspected somats consistently revealed band differences as shown in Fig. 2 and thus we consider threshold 0 to be a good indication of genotype membership.
Table 2.
Summary of genetic/genotypic data collected in neighbourhoods (NH) within three populations of G. rhizomatosa at the micro-scale of sampling every plant within a 25-m2 area
| No. of genotypes (no. of plants in each clone network) at threshold levels 0–4 |
Threshold 0 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Population | n | 0 | 1 | 2 | 3 | 4 | Private bands | P% | G/N | I | D | E |
| Cas–NH1-SI | 25 | 25 (1) | 22 (1–3) | 20 (1–4) | 17 (1–7) | 15 (1–7) | 3 | 54 | 1·00 | 0·207 | 1·00 | 0·0 |
| Cas–NH2-SI | 23 | 23 (1) | 22 (1–2) | 20 (1–3) | 16 (1–4) | 11 (1–8) | 6 | 50 | 1·00 | 0·205 | 1·00 | 0·0 |
| CasW–NH3-sterile | 56 | 43 (1–5) | 41 (1–5) | 36 (1–6) | 31 (1–11) | 15 (1–35) | 16 | 49 | 0·77 | 0·175 | 0·986 | 0·84 |
| Mull–NH4-sterile | 32 | 30 (1–2) | 29 (1–3) | 20 (1–10) | 12 (1–21) | 10 (1–22) | 1 | 34 | 0·94 | 0·149 | 0·996 | 0·52 |
| Mull–NH5-sterile | 26 | 25 (1–2) | 24 (1–2) | 21 (1–3) | 16 (1–9) | 8 (1–18) | 2 | 29 | 0·96 | 0·126 | 0·997 | 0·052 |
| Total | 162 | 146 | 138 | 117 | 92 | 59 | 28 | |||||
n = number of plants sampled.
No. of genotypes = number of different genotypes [based on thresholds of 0 (100 % matching bands) and thresholds 1–4 which indicate the maximum genetic distance that is allowed between two individuals to still be clone-mates with the same multilocus genotype]. The size of the clone network is given in parenthesis as the number of clone-mate plants.
Private bands = the number of bands unique to the population; P% = percentage of bands that were variable within populations; G/N = proportion of distinguishable genotypes; I = Shannon's Information Index; D = Simpson's Diversity Index; E = Genotypic evenness measure of Fager.
Populations: Cascade (Cas) neighbourhood 1 and 2, Cascade Walk (CasW) neighbourhood 3, Mulligan's Hut (Mull) neighbourhoods 4 and 5.
SI = plants are self-incompatible; sterile = flowers male and female sterile.
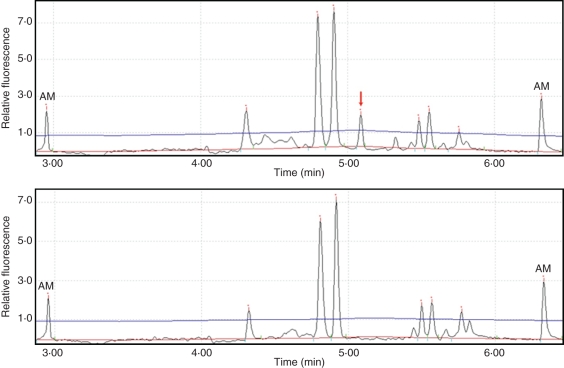
Fig. 2.
ISSR profiles of two connected individuals using primer 888. The arrow indicates a band in individual CasWk-5·1 (top) which is not present in an otherwise identical individual CasWk-5·2 (bottom).
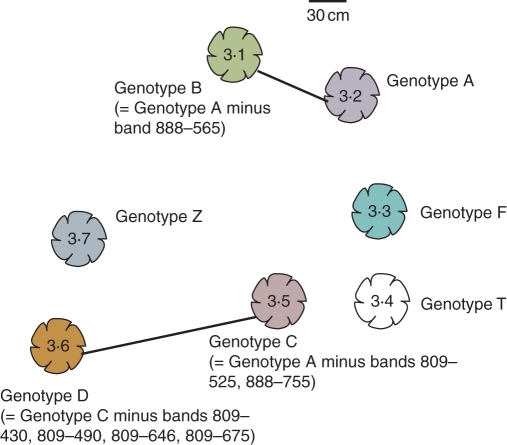
Fig. 3.
An example of a cluster of seven connected or unconnected plants and their genotypes at CascadeWalk. Plants 3·1 and 3·2 were connected below ground as were plants 3·6 and 3·5. Genotypes F–Z differed from genotype A by five to seven band differences.
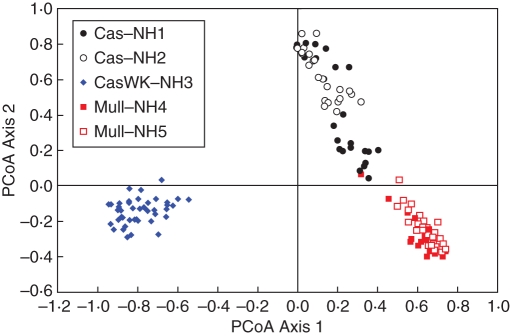
Fig. 4.
Principal co-ordinate analyses of ISSR data collected from individuals sampled from five neighbourhood populations of G. rhizomatosa in Gibraltar Range National Park, NSW (PC1 = 56·44 % variation, PC2 = 19·71 % variation, PC3 = 9·18 % variation). Populations are Cascade (Cas) neighbourhoods NH1 and NH2, Cascade Walk (CasWk) NH 3 and Mulligan's Hut (Mull) NH4 and NH5.
In summary, across neighbourhoods we found 42 instances of connected plant clusters (one or more connected ramets) and, of these, ten connected plant pairs exhibited different genotypes at the 0 threshold limit, which is unambiguous evidence of somatic mutations.
The PCoA of the neighbourhood data (Fig. 4) shows that the genetic structure within and among the five neighbourhood populations clusters strongly at the within-population level. There is minimal sharing of variation among populations with variation discretely aligned within populations. Cascade NH1 and NH2, the only populations capable of sexual reproduction in the micro-scale study, and in close proximity to each other, overlap in the distribution of several samples indicating a shared genetic history. This finding is supported by the highest values of Nei's Genetic Similarity (Table 3). The Mulligan's Hut neighbourhoods overlap on the PCoA (Fig. 3) whereas Cascade Walk is a discrete group and dissimilar to the other neighbourhoods.
Table 3.
Nei's pairwise population matrix of genetic identity for the micro-scale investigation of genetic similarity among five neighbourhoods
| Mull–NH4 | Mull–NH5 | Cas–NH1 | Cas–NH2 | CasWk–NH3 | |
|---|---|---|---|---|---|
| Mull–NH4 | 1·000 | – | – | – | – |
| Mull–NH5 | 0·985 | 1·000 | – | – | – |
| Cas–NH1 | 0·899 | 0·897 | –1·000 | – | – |
| Cas–NH2 | 0·866 | 0·859 | 0·947 | 1·000 | – |
| CasWk–NH3 | 0·780 | 0·771 | 0·850 | 0·838 | 1·000 |
1·00 indicates complete similarity.
Mull = Mulligan's Hut, Cas = Cascade, CasWk = Cascade Walk. NH = neighbourhoods 1–5.
Genotype variation: sexual recombination versus somatic mutations
The analysis of character incompatibility (Table 4 and Fig. 5A) showed that at the macro-scale, more genotypes from the fertile than the sterile populations had to be removed to produce compatibility (Fig. 5A). The self-incompatible populations contained the highest percentage of genotypes that were sexually derived (92–94 %; Table 4). The sterile populations at Mulligan's Hut and Swamp had a high proportion of sexually derived genotypes as well (79–82 %) but a greater proportion of their genotypes were derived from somatic mutations compared with the fertile populations (Table 4). In the micro-scale assessments where we sampled intensively in a 25-m2 quadrat, all of the sterile neighbourhoods had the highest percentages of genotypes derived from somatic mutagenesis (34–48 % compared with 17–20 % in self-incompatible neighbourhoods). The sterile neighbourhoods took fewer steps to resolve incompatibilities (Fig. 5B), indicating that somatic mutagenesis is a significant contributor to the origins of the genotypes in these neighbourhoods.
Table 4.
The percentage of genotypes derived from sexual recombination in five populations sampled at the macro-scale of plants no closer than 5 m and at the neighbourhood micro-scale of every plant within 25 m2
| Population | n | No. of genotypes | % of genotypes sexually derived |
|---|---|---|---|
| Macro-scale | |||
| Cascade–SI | 50 | 50 | 92 |
| Dandahra Trail–SI | 50 | 50 | 94 |
| Mulligan's Hut–sterile | 55 | 55 | 82 |
| Washpool–SC | 54 | 54 | 90 |
| Swamp–sterile | 35 | 34 | 79 |
| Micro-scale | |||
| Cascade–NH1–SI | 25 | 25 | 80 |
| Cascade–NH2–SI | 23 | 23 | 83 |
| Cascade Walk–NH3–sterile | 56 | 43 | 42 |
| Mulligan's Hut–NH4–sterile | 32 | 30 | 66 |
| Mulligan's Hut–NH5–sterile | 26 | 25 | 80 |
n = number of plants sampled.
No. of genotypes = the number of distinct ISSR profiles detected.
The percentage of genotypes sexually derived was determined from matrix incompatibility analyses as the number of genotypes that had to be removed for compatibility, divided by the total number of genotypes.
SI = plants are self-incompatible, SC = plants are self-compatible and sterile = flowers male and female sterile. NH = neighbourhoods 1–5.
Fig. 5.
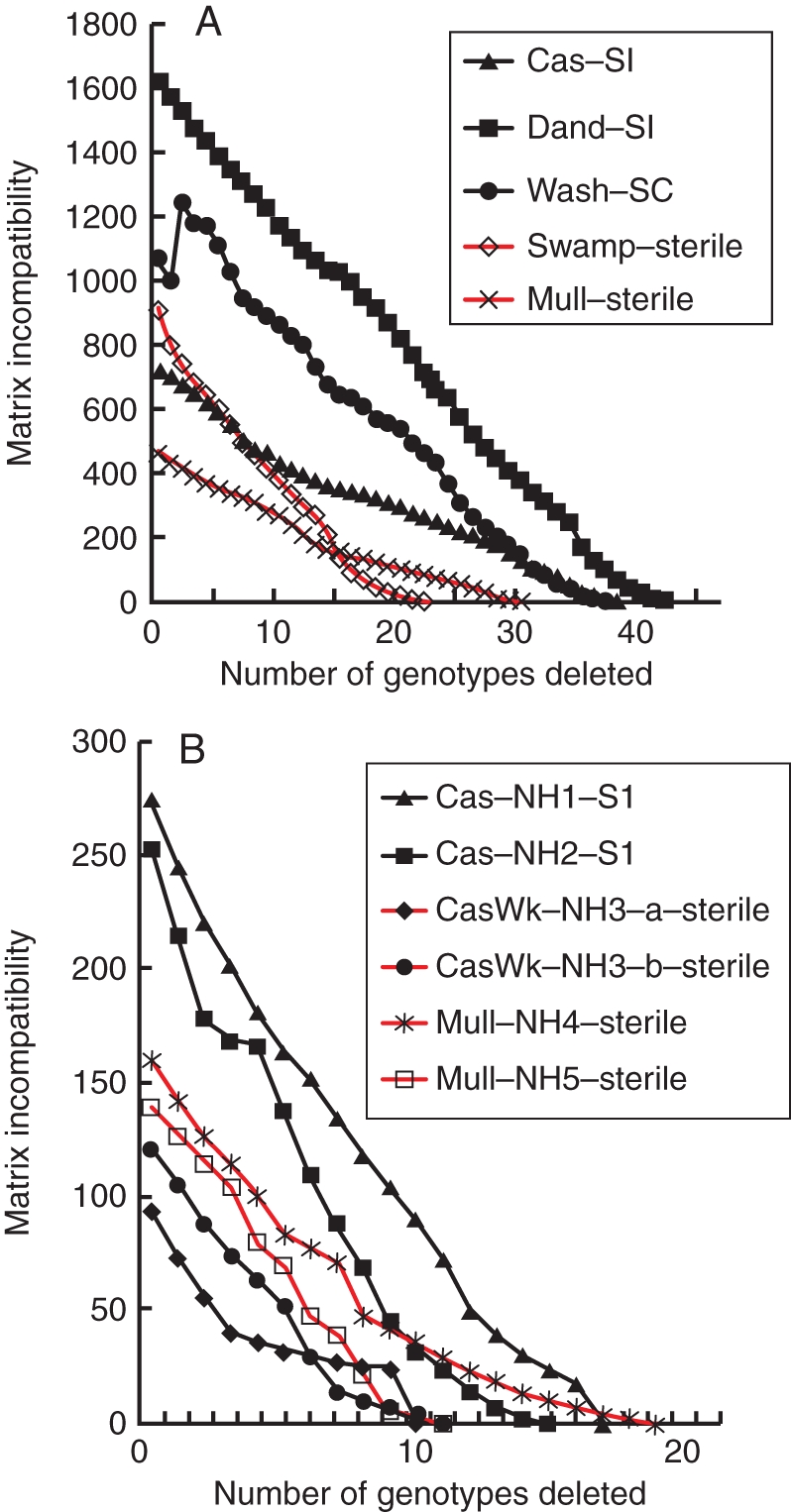
The matrix incompatibility count (MIC, the number of incompatible character states) after sequential removal of genotypes that contribute most to overall MIC at (A) for the macro-sampled populations and (B) for the micro-sampled populations. Populations are (A) Cascade, Dandahra, Mulligan's Hut (Mull), Washpool (Wash) and Swamp; (B) Cascade (Cas) neighbourhoods NH1 and NH2, Cascade Walk (CasW) NH 3, Mulligan's Hut (Mull) NH4 and NH5. For the Cascade Walk NH3 analyses, the sample was split in two to give 2 × n = 28 plants to even the sampling among neighbourhoods. SI = plants are self-incompatible; SC = plants are self-compatible; sterile = flowers male and female sterile.
DISCUSSION
Detection of somatic mutations in wild populations
The detection of somatic mutations requires variation to be revealed as unambiguous within a whole organism – whether that be a genet (Thuja plicata; O'Connell and Ritland, 2004), an agamospermous array (Taraxacum officinale; King and Schaal, 1990) or among connected ramets (Olea europaea subsp. laperrinei; Baali-Cherif and Besnard, 2005). The variable breeding systems and the semi-persistent ramets in Grevillea rhizomatosa have provided a rare opportunity to compare genetic diversity across three types of breeding systems, to investigate the origins of sterility in G. rhizomatosa and to test the hypothesis that somatic mutations contribute significantly to genetic diversity. Moderate levels of genetic diversity in G. rhizomatosa were found in both fertile (50–65 % polymorphic bands) and sterile populations (48–57 % polymorphic bands). Somatic mutations were detected in most ramet clusters that were investigated. Mutations (single- or multiple-band differences) were found on pairs of connected ramets growing in both sterile and fertile populations. In sterile Mulligan's Hut NH4, and considering all connected and unconnected plants, there were more genotypes than within the fertile populations at Cascade, although the Cascade NH1 held genotypes with 20 % more polymorphic bands than Mulligan's Hut NH4. This suggests that at Cascade sexual recombination is an important contributor to genotype diversity at this site. In the Cascade Walk neighbourhood, a sterile population with many ramet-clusters, a clone of four plants with identical genotypes was found, none of which was joined. Within this clone, members were connected to plants in seven other clusters and ramets connected in this super network mostly did not share genotypes. There were eight separate ramet clusters, suggesting that ramet connections eventually disintegrate or are disrupted giving rise to isolated plants or satellite ramet-clusters.
Genotypic diversity and breeding strategies
Our expectation that genotypic diversity would be highest in the self-incompatible populations was not supported – gene diversity was uniformly high across all populations. SC Washpool and SI Dandahra had the highest percentage of polymorphisms but genotypic diversity was equally high across all breeding strategies. The genotypic diversity found in all sterile populations was considerably higher than values reported in a meta-analysis by Honnay and Jacquemyn (2008; G/N = 0·44 ± 0·04). The same patterns of genotypic diversity were observed at the micro-scale level in which the sterile populations showed a relatively high level of variation. We suggest that somatic mutations have contributed to the high levels of observed genotypic diversity detected in this study and particularly so in SI Cascade and the sterile populations. Matrix incompatibilities showed that in all populations there are genotypes that are derived from sexual recombination. This is of particular interest in the sterile populations. In the Mull–NH5 neighbourhood for example, 80 % of genotypes had band patterns congruous with a sexual recombination event. We interpret this as persistence of initial diversity, a similar finding to that made by Kimpton et al. (2002) for Grevillea infecunda. The mutations we discovered in G. rhizomatosa using ISSR markers (i.e. neutral allele sites) are unlikely to be the mutations that induce floral sterility. Further work is required in G. rhizomatosa to investigate mutation levels in functional alleles. We hypothesize, however, that the sterility in plants is due to an accumulation of genetic damage at functional alleles within meristems (see Thomas, 2002) that has deactivated the sexual machinery, a process which can be accentuated in old and senescing plants (Ally et al., 2010). Alternative hypotheses are that rare sexual events occur in the population or that the majority of plants at this site are sterile F1 progeny from hybridization events. However, we consider it unlikely that multiple migration events involving sterile F1 progeny [of at least 25 seeds (80 % of 32 genotypes)] have occurred at Mull–NH 5, especially as nearby Mull–NH4 does not show this pattern of recombination events.
Disturbance, resprouters, sterility and somatic mutation: cause and effect?
Sterile individuals of G. rhizomatosa exhibit a collection of faulty sexual characters and processes; pollen is not distributed onto the pollen presenter prior to anthesis, pollen is mostly unviable, and viable pollen from fecund populations does not instigate seed set in the sterile Mulligan's Hut population (Gross and Caddy, 2006); i.e. sterile populations of G. rhizomatosa are both male and female sterile. Ecological factors that reduce the frequency of sexual events (e.g. flowering) and thus reduce the opportunities for successful sexual reproduction are likely to prevent the selection of sexual characters, resulting in an accumulation of mutations in sexual traits (Klekowski, 1997). This is the ‘Use it or lose it’ hypothesis discussed in Eckert (2002). The primary ecological factor in the study area that could retard flowering, alter germination success and promote clonal reproduction is a disrupted fire regime (more frequent and or severe fires). Fire is a known stimulator of clonal reproduction in some Australian ecosystems (e.g. Lacey, 1974), and fire is a significant ecological factor in the heath habitat of G. rhizomatosa (Croft et al., 2006). In 2006 much of the habitat of G. rhizomatosa had experienced two fires in the previous 10 years (Clarke et al., 2006). Croft et al. (2006) note that a fire-free interval of 10 years would be needed to ensure the persistence of G. rhizomatosa. Frequent fire episodes may prevent sexual reproduction by killing juveniles (and seed reserves) before sexual maturity (Keith, 1996), by depleting starch reserves in standing plants (Keith, 1996) or by preventing resprouting adults from flowering every year. At Washpool our field records for G. rhizomatosa show that fire destroys the above-ground biomass but plants quickly resprout from underground suckers. After the 2002 fires it took 8 years for re-sprouting plants at Washpool to recommence flowering (C. L. Gross and P. A. Nelson, unpub. res.). Frequent fires may select for the resprouting habit which allows individuals to vegetatively proliferate, thereby excluding opportunities for seedling recruitment. As summarized by Lloret et al. (1999), however, vegetative reproduction is common in many phylogenetic lineages and biogeographical contexts not necessarily associated with fire (e.g. hurricanes). In grasslands, for example, vegetative reproduction can be promoted through management regimes (e.g. Schaal and Leverich, 1996) and this has been linked to the accumulation of somatic mutations in meadows (Warren, 2009). Thus our results may be relevant to other biogeographical contexts. In this study we have shown that the mutations occur in ramets that are resprouting. Lamont and Wiens (2003) predicted that frequent axillary branching (induced by trauma) via abundant stored buds within resprouters might favour expression among their genets and ramets of beneficial somatic mutations present in their meristematic tissues. We have no evidence that the mutations we detected are those directly responsible for the observed declines in sexual fitness in G. rhizomatosa – only that somatic mutations are present. Moreover, the effects of the somatic mutations may be largely masked in the diploid adult phase but be deleterious in the haploid pollen stage, causing pollen unviability. We also do not know whether the mutations we detected are beneficial to G. rhizomatosa; i.e. at the expense of sexual reproduction, vegetative reproduction may be enhanced (antagonistic pleiotropy; see Dorken et al., 2004). As the production of somatic mutations is likely to be amplified in G. rhizomatosa, a long-lived species, a high mutation rate may also result in the loss of functional traits (Muller, 1964). Selection for optimization of mutation rates has been proposed for asexual organisms and, as pointed out by King and Schaal (1990), the processes that generate genetic variation may themselves be under selection. As sexual functionality has been lost in several populations of G. rhizomatosa, it would be extremely informative to investigate the functional relationship between sterility and genetic diversity in this species. Sterility in populations of G. rhizomatosa was not commensurate with a complete lack of genotypic diversity. The Mulligan's Hut and Swamp populations had relatively high levels of diversity when compared with fertile populations.
The detection of high genotype variability in sterile populations suggests several scenarios: (a) sterile populations were founded by fertile genotypes and sterility has subsequently developed; (b) sterile populations originated from genetically variable but sterile individuals, e.g. F1 hybrids; or (c) that populations consist of large clones in which somatic mutation has generated genetic variability but this variability has resulted in a loss of traits. These hypotheses are not mutually exclusive. Interestingly, the two sterile populations are genetically distinct from one another. Swamp is more similar to SC Washpool than sterile Mulligan's Hut. This suggests that each population may have been founded independently and that different pathways for sterility may be involved.
Multiple breeding systems in Grevillea rhizomatosa
Smith and Gross (2002, table 1) collated data for 21 species of Grevillea and 19 of these were self-compatible. Further assessments of breeding systems in Grevillea have revealed additional self-compatible species (G. repens; Holmes et al., 2008), although some weakly so [G. iaspicula; fig. 2 in Hoebee and Young, 2001 (however, illegal access to pollinator exclusion bags by honeybees bringing in outcross pollen may have been involved here; S. Hoebee, pers. comm., 10 June 2010)] and fully sterile species (G. infecunda; Kimpton et al., 2002). Interestingly, for two species (G. robusta and G. sphacelata) both self-compatible and self-incompatible populations have been detected [G. robusta (Brough, 1933; Kalinganire et al., 2000); G. sphacelata (Hermanutz et al., 1998; Richardson et al., 2000)]. For G. robusta the incompatibility mechanism has been determined to be gametophytic self-incompatibility (Kalinganire et al., 2000). In G. rhizomatosa, plants at Washpool are self-compatible but at Cascade and Dandahra plants are self-incompatible. Here, we hypothesize that selfing is a synapomorphic character for Grevillea and that self-incompatibility is imposed through inbreeding depression on G. rhizomatosa at Cascade and Dandahra where selfing has brought together a high load of lethal mutations. Inbreeding depression is elevated in perennials with high mitotic mutation rates (Morgan, 2001) and Barrett et al. (1996) suggest that the evolution of selfing would be constrained by high genetic loads. We hypothesize that the populations at Cascade and Dandahra have high genetic loads resulting in strong inbreeding depression. The self-incompatible populations (Cascade and Dandahra) had the highest occurrence of polymorphic bands, and somatic mutations were also detected in Cascade (Dandahra not tested). We suggest that a high level of polymorphism in these populations is evidence of a high mutational load rather than evidence of superior genetic health. At Mulligan's Hut, Swamp and Cascade Walk, we propose that the mutational load has passed a threshold resulting in dysfunctional reproductive characters (faulty anthesis, inviable pollen, female sterility). Support for this hypothesis can be found in Gross and Caddy (2006, fig. 4) where high levels of unviable pollen were detected at Cascade and Dandahra compared with Washpool, suggesting that populations at Cascade and Dandahra may be moving towards a mutational meltdown. Finally, we propose that the mutational meltdown that induces sterility in G. rhizomatosa is unable to be rectified through selection against the deleterious mutations at the cellular level because the proliferation of the clones is actively selected in the fire-prone landscape where flowering can be retarded after fire.
Clonality in context and the Australian flora
Clonal growth can afford persistence in harsh environments (Song et al., 2002). However, while the importance of clonal behaviour in plants is appreciated for many floras (e.g. central Europe, Klimeš and Klimešová, 1999; north-east China, Song et al., 2002), the rich Australian flora (approx. 20 000 angiosperms) has not yet been evaluated from this perspective at a meta-analysis level (e.g. Sjöström and Gross, 2006; Gross, 2005). In a continent that is subjected to extreme environmental disturbances and conditions (e.g. drought, floods, fire, cyclones, infertile soils) the clonal strategy in Australian ecosystems is potentially prevalent and very important for population persistence during such times where flowering and fruiting is arrested or prevented. There are some well-recognized and significant occurrences of facultative clonal growth patterns among the Australian flora that occupy ecosystems dependent upon disturbance (e.g. Triodia; Lazarides, 1997) and within the alien flora there are many that flourish in disturbed habitats [e.g. facultatively clonal Phyla canescens (Xu et al., 2010; Price et al., 2011); obligately clonal Arundo donax (A. Haddadchi et al., unpubl. res.)]. Increasingly, covert clonality is being discovered in the Australian endangered tree flora [e.g. Elaeocarpus williamsianus (Rossetto et al., 2004); Wollemia nobilis (Peakall et al., 2003)] that is most certainly associated with persistence under adversity [see also Erythroxylum pusillum (van der Merwe et al., 2010)]. Within certain shrubby and herbaceous flora clonality appears to be important too and may provide assurance during other adverse situations such as a shortage of pollinators [Canavalia rosea (Gross, 1993); Tetratheca juncea (Gross et al., 2003)] or in marginal environments (Banksia ionthocarpa ssp. chrysophoenix; Millar et al., 2010). There is thus much to be learnt about the benefits and extent of clonal reproduction in the Australian flora. Our work has shown that genotypic diversity in a clonal species that is endemic to a fire-prone landscape is derived from both recombination and somatic mutagenesis and the latter is particular prevalent in sterile populations. The compensation that somatic mutagenesis may provide for sterile (and fertile) populations in lieu of the benefits from recombination is generally unknown. We suggest that, as a starting point, partitioning the origins of genetic diversity alongside the elucidation of clonality will contribute very important information on the evolution, speciation and persistence ecology of the Australian flora.
Conclusions
Asexual reproduction allows Grevillea rhizomatosa to persist in a landscape where fecundity is likely to be retarded by frequent fires. However, a lack of sexual recombination in some populations has not diminished genotype diversity and we have shown here that diversity is generated through the advent of somatic mutations. Tuskan et al. (1996, and references therein) suggest that somatic mutations provide clonal organisms with a mechanism for enhanced survival and fitness, particularly for long-lived organisms that occupy variable environments. We concur with this view but add that the mutational increase in somatic mutations may have been at the expense of successful sexual reproduction. Further work is required to determine the ultimate causes of sterility in G. rhizomatosa, and whether a build-up of somatic mutations in apical meristems handicaps the sexual machinery. From a conservation perspective, management regimes should aim to minimize disruption to sexual processes, a theme emphasized by Honnay and Bossuyt (2005).
ACKNOWLEDGEMENTS
We thank D. Mackay for field assistance, B. Makinson for information on Grevillea, H. Caddy for field information and L. Kumar for fire information. E. James and S. Hoebee provided valuable comments on an earlier version of this paper. Our work benefited from discussions with J. Bruhl, D. Drake and B. Lamont. The students of the 2008 class of Conservation Biology (UNE) are thanked for helping with mapping the Cascade Walk neighbourhood. This work was conducted under NPWS permit NZ143 and was funded by UNE.
LITERATURE CITED
- Ally D, Ritland K, Otto SP. Can clone size serve as a proxy for clone age? An exploration using microsatellite divergence in Populus tremuloides. Molecular Ecology. 2008;17:4897–4911. doi: 10.1111/j.1365-294X.2008.03962.x. [DOI] [PubMed] [Google Scholar]
- Ally D, Ritland K, Otto SP. Aging in a long-lived clonal tree. PLoS Biology. 2010;8 doi: 10.1371/journal.pbio.1000454. e1000454. http://dx.doi.org/10.1371/journal.pbio.1000454 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baali-Cherif D, Besnard G. High genetic diversity and clonal growth in relict populations of Olea europaea subsp. laperrinei (Oleaceae) from Hoggar, Algeria. Annals of Botany. 2005;96:823–830. doi: 10.1093/aob/mci232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SCH, Harder LD, Worley AC. The comparative biology of pollination and mating in flowering plants. Philosophical Transactions of the Royal Society of London Series B – Biological Sciences. 1996;351:1271–1280. [Google Scholar]
- Brough P. The life history of Grevillea robusta. Proceedings of the Linnean Society of New South Wales. 1933;58:33–73. [Google Scholar]
- Caddy HAR, Gross CL. Population structure and fecundity in the putative sterile shrub, Grevillea rhizomatosa Olde & Marriott (Proteaceae) Proceedings of the Linnean Society of New South Wales. 2006;127:11–18. [Google Scholar]
- Chaiwongsar S, Otegui MS, Jester PJ, Monson SS, Krysan PJ. The protein kinase genes MAP3Ke1 and MAP3Ke2 are required for pollen viability in Arabidopsis thaliana. The Plant Journal. 2006;48:193–205. doi: 10.1111/j.1365-313X.2006.02863.x. [DOI] [PubMed] [Google Scholar]
- Clarke PJ, Coughlin R, Kumar L. Fire history and fire regimes: implications for plant conservation in New England Tableland National Parks. Armidale, NSW, Australia: University of New England; 2006. In: Unpublished Report for DECC. [Google Scholar]
- Croft P, Hofmeyer D, Hunter JT. Fire responses in four rare plant species at Gibraltar Range National Park, Northern Tablelands, NSW. Proceedings of the Linnean Society of New South Wales. 2006;127:57–62. [Google Scholar]
- Dorken ME, Neville KJ, Eckert CG. Evolutionary vestigialization of sex in a clonal plant: selection versus neutral mutation in geographically peripheral populations. Proceedings of the Royal Society B. 2004;271:2375–2380. doi: 10.1098/rspb.2004.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert CG. The loss of sex in clonal plants. Evolutionary Ecology. 2002;15:501–520. [Google Scholar]
- Eckert CG, Dorken ME, Mitchell SA. Loss of sex in clonal populations of a flowering plant, Decodon verticillatus (Lythraceae) Evolution. 1999;53:1079–1092. doi: 10.1111/j.1558-5646.1999.tb04523.x. [DOI] [PubMed] [Google Scholar]
- Ellstrand NC, Roose ML. Patterns of genotypic diversity in clonal plant species. American Journal of Botany. 1987;74:123–131. [Google Scholar]
- Fager EW. Diversity: a sampling study. The American Naturalist. 1972;106:293–310. [Google Scholar]
- Fatemi M, Gross CL. Life on the edge: high levels of genetic diversity in a cliff population of Bertya ingramii are attributed to B. rosmarinifolia (Euphorbiaceae) Biological Conservation. 2009;142:1461–1468. [Google Scholar]
- Felsenstein J. The evolutionary advantage of recombination. Genetics. 1974;78:737–756. doi: 10.1093/genetics/78.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin ID, Aitken EAB, Smith LW. Application of inter simple sequence repeat (ISSR) markers to plant genetics. Electrophoresis. 1997;18:1524–1528. doi: 10.1002/elps.1150180906. [DOI] [PubMed] [Google Scholar]
- Gross CL. The reproductive ecology of Canavalia rosea (Fabaceae) on Anak Krakatau, Indonesia. Australian Journal of Botany. 1993;41:591–599. [Google Scholar]
- Gross CL. A comparison of the sexual systems in the trees from the Australian tropics with other tropical biomes: more monoecy but why? American Journal of Botany. 2005;92:907–919. doi: 10.3732/ajb.92.6.907. [DOI] [PubMed] [Google Scholar]
- Gross CL, Caddy HAR. Are differences in breeding mechanisms and fertility among populations contributing to rarity in Grevillea rhizomatosa (Proteaceae)? American Journal of Botany. 2006;93:1791–1799. doi: 10.3732/ajb.93.12.1791. [DOI] [PubMed] [Google Scholar]
- Gross CL, Bartier FV, Mulligan DR. Floral structure, breeding system and fruit-set in the threatened sub-shrub Tetratheca juncea Smith (Tremandraceae) Annals of Botany. 2003;92:771–777. doi: 10.1093/aob/mcg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanutz L, Innes D, Denham A, Whelan R. Very low fruit:flower ratios in Grevillea (Proteaceae) are independent of breeding system. Australian Journal of Botany. 1998;46:465–478. [Google Scholar]
- Hoebee SE, Young AG. Low neighbourhood size and high interpopulation differentiation in the endangered shrub Grevillea iaspicula McGill (Proteaceae) Heredity. 2001;86:489–496. doi: 10.1046/j.1365-2540.2001.00857.x. [DOI] [PubMed] [Google Scholar]
- Holmes GD, James EA, Hoffmann AA. Limitations to reproductive output and genetic rescue in populations of the rare shrub Grevillea repens (Proteaceae) Annals of Botany. 2008;102:1031–1041. doi: 10.1093/aob/mcn195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes GD, James EA, Hoffmann AA. Divergent levels of genetic variation and ploidy among populations of the rare shrub, Grevillea repens (Proteaceae) Conservation Genetics. 2009;10:827–837. [Google Scholar]
- Honnay O, Bossuyt B. Prolonged clonal growth: escape route or route to extinction? Oikos. 2005;108:427–432. [Google Scholar]
- Honnay O, Jacquemyn H. A meta-analysis of the relation between mating system, growth form and genotypic diversity in clonal plant species. Evolutionary Ecology. 2008;22:299–312. [Google Scholar]
- Kalinganire A, Harwood CE, Slee MU, Simons AJ. Floral structure, stigma receptivity and pollen viability in relation to protandry and self-incompatibility in silky oak (Grevillea robusta A. Cunn.) Annals of Botany. 2000;86:133–148. [Google Scholar]
- Keith DA. Fire-driven extinction of plant populations: a synthesis of theory and review of evidence from Australian vegetation. Proceedings of the Linnean Society of New South Wales. 1996;116:37–78. [Google Scholar]
- Kimpton SK, James EA, Drinnan AN. Reproductive biology and genetic marker diversity in Grevillea infecunda (Proteaceae), a rare plant with no known seed production. Australian Systematic Botany. 2002;15:485–492. [Google Scholar]
- King LM, Schaal BA. Genotypic variation within asexual lineages of Taraxacum officinale. Proceedings of the National Academy of Sciences of the USA. 1990;87:998–1002. doi: 10.1073/pnas.87.3.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klekowski EJ. Somatic mutation theory of clonality. In: de Kroon H, van Groenendael J, editors. The ecology and evolution of clonal growth in plants. Leiden, The Netherlands: Backhuys Publishers; 1997. pp. 227–241. [Google Scholar]
- Klimeš L, Klimešová J. CLO-PLA2 – a database of clonal plants in central Europe. Plant Ecology. 1999;141:9–19. [Google Scholar]
- Lacey CJ. Rhizomes in tropical eucalypts and their role in recovery from fire damage. Australian Journal of Botany. 1974;22:29–38. [Google Scholar]
- Lamont BB, Wiens D. Are seed set and speciation rates always low among species that resprout after fire, and why? Evolutionary Ecology. 2003;17:277–292. [Google Scholar]
- Lazarides M. A revision of Triodia including Plectrachne (Poaceae, Eragrostideae, Triodiinae) Australian Systematic Botany. 1997;10:381–489. [Google Scholar]
- Lloret F, Verdú M, Flores-Hernández N, Valiente-Banuet A. Fire and resprouting in Mediterranean ecosystems: insights from an external biogeographical region in the Mexical shrubland. American Journal of Botany. 1999;86:1655–1661. [PubMed] [Google Scholar]
- Lo EYY, Stefanovic S, Ritland K, Dickinson TA. Fine scale comparisons of genetic variability in seed families of asexually and sexually reproducing Crataegus (Hawthorn; Rosaceae) American Journal of Botany. 2010;97:1014–1024. doi: 10.3732/ajb.0900091. [DOI] [PubMed] [Google Scholar]
- Lynch AJJ, Barnes RW, Cambecedes J, Vaillancourt RE. Genetic evidence that Lomatia tasmanica (Proteaceae) is an ancient clone. Australian Journal of Botany. 1998;46:25–33. [Google Scholar]
- Meirmans PG, Van Tienderen PH. GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Molecular Ecology Notes. 2004;4:792–794. [Google Scholar]
- van der Merwe M, Spain CS, Rossetto M. Enhancing the survival and expansion potential of a founder population through clonality. New Phytologist. 2010;188:868–878. doi: 10.1111/j.1469-8137.2010.03396.x. [DOI] [PubMed] [Google Scholar]
- Mes THM. Character compatibility of molecular markers to distinguish asexual and sexual reproduction. Molecular Ecology. 1998;7:1719–1727. [Google Scholar]
- Millar MA, Byrne M, Coates DJ. The maintenance of disparate levels of clonality, genetic diversity and genetic differentiation in disjunct subspecies of the rare Banksia ionthocarpa. Molecular Ecology. 2010;19:4217–4227. doi: 10.1111/j.1365-294X.2010.04817.x. [DOI] [PubMed] [Google Scholar]
- Morgan MT. Consequences of life history for inbreeding depression and mating system evolution in plants. Proceedings of the Royal Society of London Series B – Biological Sciences. 2001;268:1817–1824. doi: 10.1098/rspb.2001.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ. The relation of recombination to mutational advantage. Mutational Research. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- Neal JM, Hardner CM, Gross CL. Population demography and fecundity do not decline with habitat fragmentation in the rainforest tree Macadamia integrifolia (Proteaceae) Biological Conservation. 2010;143:2591–2600. [Google Scholar]
- Nelson PA. 2006. The impact of reproductive divergence and clonality on the genetic diversity of the threatened species Grevillea rhizomatosa Olde & Marriot (Proteaceae). MNatRes Thesis, University of New England, Armidale, NSW, Australia. [Google Scholar]
- O'Connell LM, Ritland K. Somatic mutations at microsatellite loci in western red cedar (Thuja plicata: Cupressaceae) Journal of Heredity. 2004;95:172–176. doi: 10.1093/jhered/esh024. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Ebert D, Scott LJ, Meagher PF, Offord CA. Comparative genetic study confirms exceptionally low genetic variation in the ancient and endangered relictual conifer, Wollemia nobilis (Araucariaceae) Molecular Ecology. 2003;12:2331–2343. doi: 10.1046/j.1365-294x.2003.01926.x. [DOI] [PubMed] [Google Scholar]
- Pielou EC. An introduction to mathematical ecology. New York, NY: Wiley-Interscience; 1969. [Google Scholar]
- Pisanu PC, Gross CL, Flood L. Reproduction in wild populations of the threatened tree Macadamia tetraphylla: interpopulation pollen enriches fecundity in a declining species. Biotropica. 2009;41:391–398. [Google Scholar]
- Price JN, Macdonald MJ, Gross CL, Whalley RDB, Simpson IH. Vegetative reproduction facilitates early expansion of Phyla canescens in a semi-arid floodplain. Biological Invasions. 2011;13:285–289. [Google Scholar]
- Pusterla N, Mapes S, Rejmanek D, Gebhart C. Detection of Lawsonia intracellularis by real-time PCR in the feces of free-living animals from equine farms with documented occurrence of equine proliferative enteropathy. Journal of Wildlife Diseases. 2008;44:992–998. doi: 10.7589/0090-3558-44.4.992. [DOI] [PubMed] [Google Scholar]
- Richardson MBG, Ayre DJ, Whelan RJ. Pollinator behaviour, mate choice and the realised mating systems of Grevillea mucronulata and Grevillea sphacelata. Australian Journal of Botany. 2000;48:357–366. [Google Scholar]
- Rossetto M, Gross CL, Jones R, Hunter J. The impact of clonality on an endangered tree (Elaeocarpus williamsianus) in a fragmented rainforest. Biological Conservation. 2004;117:33–39. [Google Scholar]
- Schaal BA, Leverich WJ. Molecular variation in isolated plant populations. Plant Species Biology. 1996;11:33–40. [Google Scholar]
- Scott B, Gross CL. Recovery directions for monoecious and endangered Bertya ingramii using autecology and comparisons with common B. rosmarinifolia (Euphorbiaceae) Biodiversity and Conservation. 2004;13:885–899. [Google Scholar]
- Sjöström A, Gross CL. Life-history characters and phylogeny are correlated with extinction risk in the Australian angiosperms. Journal of Biogeography. 2006;33:271–290. [Google Scholar]
- Smith JA. 2004 Reproductive, developmental and genetic factors associated with sterility in the endangered, clonal shrub Hakea pulvinifera L.A.S.Johnson (Proteaceae). PhD Thesis University of New England, New South Wales, Australia. [Google Scholar]
- Smith JA, Gross CL. The pollination ecology of Grevillea beadleana McGillivray, an endangered shrub from northern New South Wales, Australia. Annals of Botany. 2002;89:97–108. doi: 10.1093/aob/mcf015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, Dong M, Jiang G. Importance of clonal plants and plant species diversity in the Northeast China Transect. Ecological Research. 2002;17:705–716. [Google Scholar]
- Thomas H. Ageing in plants. Mechanisms of Ageing and Development. 2002;123:747–753. doi: 10.1016/s0047-6374(01)00420-1. [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Francis KE, Russ SL, Romme WH, Turner MG. RAPD markers reveal diversity within and among clonal and seedling stands of aspen in Yellowstone National Park, USA. Canadian Journal of Forest Research – Revue Canadienne de Recherche Forestiere. 1996;26:2088–2098. [Google Scholar]
- Vanormelingen P, Verleyen E, Vyverman W. The diversity and distribution of diatoms: from cosmopolitanism to narrow endemism. Biodiversity and Conservation. 2008;17:393–405. [Google Scholar]
- Warren J. Extra petals in the buttercup (Ranunculus repens) provide a quick method to estimate the age of meadows. Annals of Botany. 2009;104:785–788. doi: 10.1093/aob/mcp139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M. PICA 4·0: software and documentation. London: Department of Zoology, The Natural History Museum; 2001. [Google Scholar]
- de Witte LC, Stöcklin J. Longevity of clonal plants: why it matters and how to measure it. Annals of Botany. 2010;106:849–857. doi: 10.1093/aob/mcq191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AD, Xiang QY, Kephart SR. Assessing hybridization in natural populations of Penstemon (Scrophulariaceae) using hypervariable intersimple sequence repeat (ISSR) bands. Molecular Ecology. 1998;7:1107–1125. doi: 10.1046/j.1365-294x.1998.00425.x. [DOI] [PubMed] [Google Scholar]
- Xu CY, Julien MH, Fatemi M, et al. Phenotypic divergence during the invasion of Phyla canescens in Australia and France: evidence for selection-driven evolution. Ecology Letters. 2010;13:32–44. doi: 10.1111/j.1461-0248.2009.01395.x. [DOI] [PubMed] [Google Scholar]