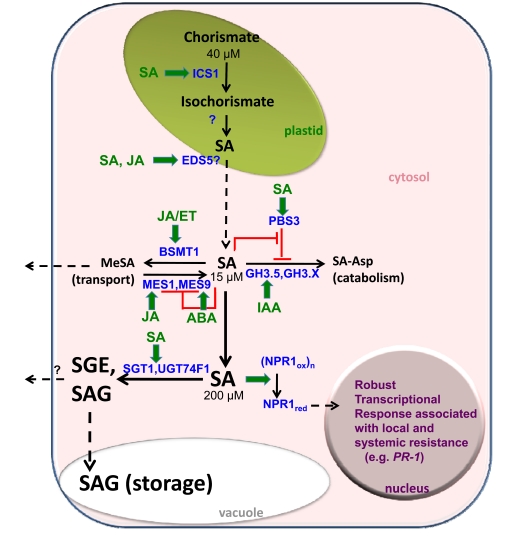
Figure 3.
Model integrating current knowledge of biochemical and transcriptional regulation of SA synthesis, SA modification, and activation of robust defense-associated responses via reduced, nuclear-localized NPR1.
SA-dependent NPR1-independent processes are not indicated here. Although all processes are shown in one cell, it is likely that spatial and temporal separation of responses is critical to their function. Dashed lines indicate transport across membrane(s). Concentrations shown reflect the Km, of the enzymes for SA. Feedback inhibition of enzyme activity is shown in red. Hormone-induced gene expression is indicated by thick green arrows, based on data from AtGenExpress Hormone and Chemical data series, Exogenous SA treatment of Whole Leaves experiment (NCBI Gene Expression Omnibus Acc. No. GSE33402), and the literature (in text). Transcription factors regulating gene activity are not shown. Although several other AtMES enzymes are active on SA, only the two most active are shown for brevity. Abbreviations are as follows: abscisic acid (ABA), benzoic acid/ salicylic acid carboxyl methyltransferase 1 (BSMT1), enhanced disease susceptibility 5 (EDS5), ethylene (ET), known and/or unknown members of the GH3 acyl adenylase family (GH3.5/GH3.X), indole-3-acetic acid (IAA), isochorismate synthase 1 (ICS1), jasmonic acid (JA), methyl esterase 1 and 9 (MES1/9), methyl salicylate (MeSA), oxidized or reduced forms of Non-expressor of Pathogenesis-Related genes 1 (NPR1ox/NPR1red), avrP-phB susceptible 3 (PBS3), pathogenesis-related 1 gene (PR-1), salicylic acid (SA), salicyloyl-L-aspartate (SA-Asp), SA 2-O-β-D-glucoside (SAG), salicylate glucose ester (SGE), salicylic acid glucosyltransferase 1 (SGT1), UDP-glucosyltransferase 74F1 (UGT74F1). See text for details and supporting information. Adapted from Okrent et al. (2011).