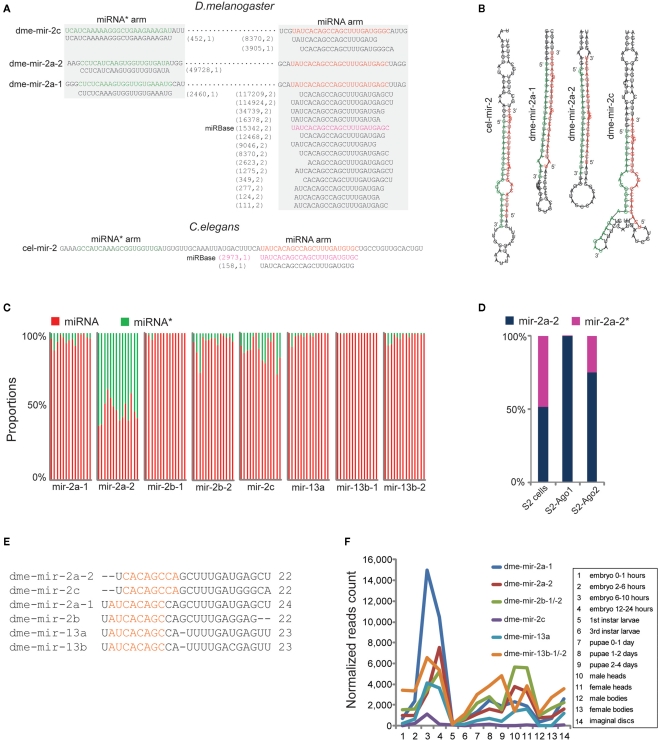
Figure 3.
Correction of dme-miR-2 family needs manual inspection of their miRNA/miRNA* duplexes. (A) The dme-mir-2a-1, dme-mir-2a-2, and dme-mir-c possess identical miRNA arms, on which the smRNA-seq reads were multiply mapped. The two numbers in the brackets are the count of smRNA-seq reads and the count of mapped locations in the genome for each isoform. (B) For the hairpin structures, we found that the 24-nt mir-2a isoform paired with mir-2a-1*, the 22-nt mir-2a isoform paired with mir-2a-2*, and 22-nt mir-2c isoform paired with mir-2c* to form the correct miRNA/miRNA* duplexes with 2-nt overhang at 3′ ends. (C) The proportions of the corrected miRNA and miRNA* strands for each miR-2 member across 14 samples. Each column represents one sample. The mir-2a-2 is the only exception that have equal amount of miRNA and miRNA* strands across all the samples. (D) Even mir-2a-2 and mir-2a-2* both existed in S2 cells, only the guide strands were associated with Ago proteins. (E) The alignment of corrected miR-2 family members. (F) The rebuilt developmental expression profile of D. melanogaster miR-2 family using the corrected miR-2 family sequences.