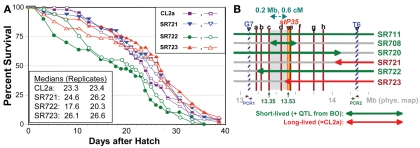
Figure 6.
Second-round screen for lsq4 recombinants. (A) Survivals of C. elegans wild-type strain CL2a and three recombinant-congenic lines derived from SR711 (see Figure 4) during three further generations of backcross to CL2a. Recombinants were identified and endpoints mapped as described for the initial screening cycle (see Figure 4). (B) Map based on survival data [(A) and Figure 4], and fine-mapping of endpoints for introgressed (BO-derived) segments. In addition to the three recombinant-congenic lines assessed in (A), (B) summarizes fine-mapping and survival data for lines SR711, SR708, and SR720 (Figure 4 and data not shown). Red lines a–h indicate positions of eight informative SNPs mapped within the G7 – T6 interval; b–h correspond to SNP triangles in Figure 4C). Lsq4 lies to the right of “c” (because SR708 = CL2a at that marker, but carries the BO/shorter life-span trait of lsq4), and left of stP35 (because SR723 = BO there but carries the CL2a/longer life-span trait). PCR1 and PCR2 indicate polymerase chain reaction assays used to genotype flanking markers G7 and T6, respectively, to ascertain recombinants in sublines. PCR1 primers: F, 5′-ACTCTTCGAAGACAACTC-3′; R, 5′-TCTTCAGAATGCTCCGCC-3′. PCR2 primers: F, 5′-TTTGGAGAAGGGTGTAATGC-3′; R, 5′-GGAAGATTTGGAGAAGGGTG-3′.