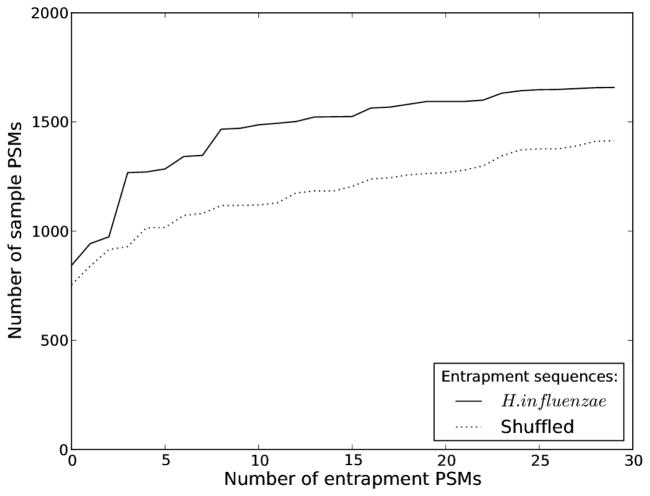
Figure 1. The dependency on entrapment sequences.
Spectra derived from a known protein mixture were scored against bipartite databases consisting of the known sample sequences and two different compositions of entrapment sequences. From the resulting PSMs we plotted the number of sample PSMs accepted for increasing numbers of entrapment PSMs, considering their XCorr score. The two different entrapment versions of the database had the same number of peptides, and sequences were either H. influenzae or shuffled versions of the sample proteins.