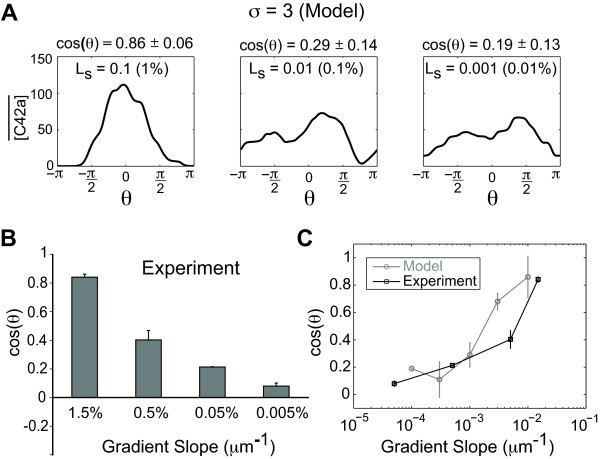
Figure 5.
Directional accuracy in yeast model simulations and microfluidics experiments. (A) Simulations of yeast model at different gradient slopes. Lslp = 0.1, 0.01, and 0.001 nM μm-1 (Lmid = 10 nM) and σ = 3. Using the wild-type model, at least 20 Monte Carlo simulations were performed as described above. The mean value of active Cdc42 is plotted, and the mean ± SEM of cos(θ) is also shown for each slope. (B) Directional projection accuracy at different gradient slopes. Experiments were performed for 1.5% gradient, 0.5%, 0.05%, and 0.005% μm-1 gradients (Lslp/Lmid). Cells were counted in the middle two sections of the gradient where Lmid ~ 20 nM. Directional accuracy was measured in terms of cos(θ), and the mean ± SEM is shown for n = 3 trials. (C) Plotting yeast mating projection directional accuracy as a function of gradient slope for both experiments and modeling. Simulation data (gray) is from (A) and experimental data (black) is from (B).