Figure 1. Phosphorylation of S463 within the CRN2 coiled coil does not induce trimer disassembly.
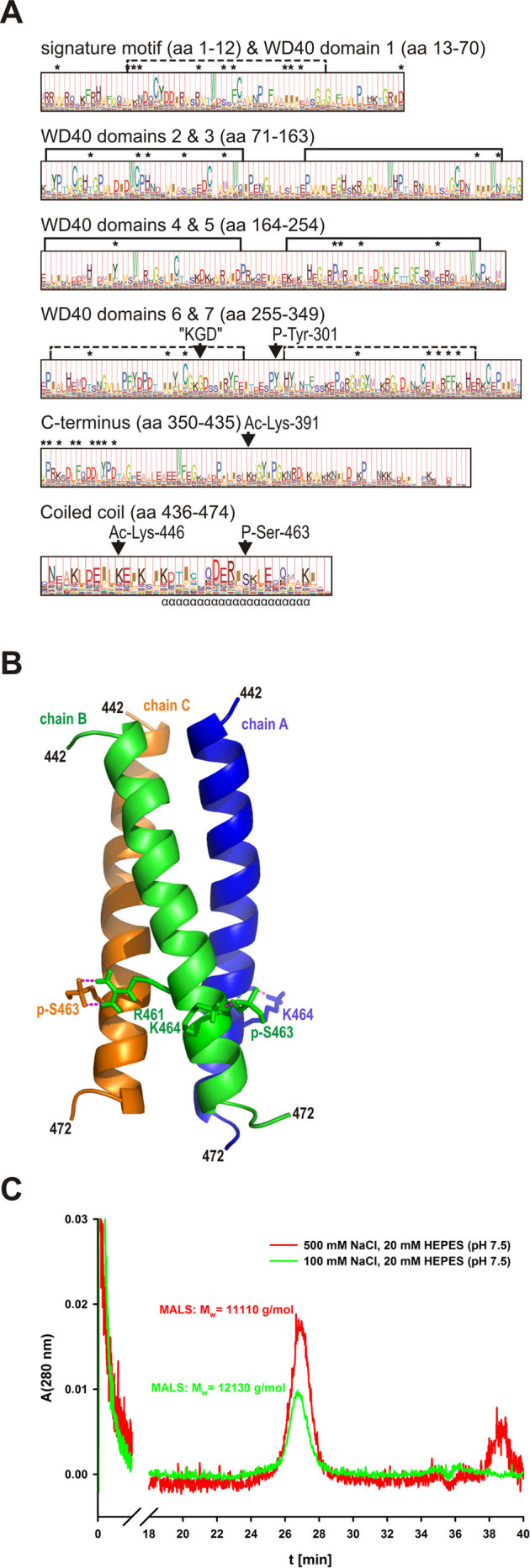
(A) profile hidden Markov model (pHMM) of the CRN2 subfamily. The probability distribution of amino acids within the CRN2 subfamily is reflected by letter height while the “functional significance” predicted by HMMER is given by the full column height at each site. The Ser-463 phosphorylation site is uniquely conserved within the CRN2 subfamily, whereas sites of other predicted post-translational changes (P-Tyr-301, Ac-Lys-391 and Ac-Lys-446) are common to a limited number of other coronin subfamilies. Asterisks mark amino acids identified by SDPfox which show evidence of a conservation pattern able to distinguish individual coronin subfamilies and are therefore taken to confer “functional specificity”. These SDPs localized mainly to regions of the N- and C-terminal domains, in contrast to the “KGD” motif universally conserved in coronin proteins (aa485-487 in CRN2). (B) homology model of the trimeric CRN2 coiled coil (aa442-447, monomer chains A, B, C) based on the crystal structure of the trimeric coiled coil of hemagglutinin (HA2 chain) using residues 74 to 113 from chain B (36; PDB 1eo8). Interactions of phosphorylated S463 (pS463, see C) with R461 and K464 are illustrated. (C) size-exclusion chromatogram of synthetic pS463-CRN2 peptide (18 mg/ml) acquired on a Superose-12 column under normal (100 mM) and high salt (500 mM NaCl) conditions. The peptide eluted as single species whose molecular mass was determined using the online multi-angle light scattering (MALS) detector; theoretical mass of the trimer is 13,959 g/mol.
