Table 3. Performance comparisons for eukaryotic protein subcellular location prediction method based on the iLoc8897 dataset.
| Subcellular location | Euk-mPLoc 2.0 (2010) (Chou and Shen 2010) | iLoc-Euk (2011) (Chou et al. 2011) | LIBSVM | KNN | The proposed method | |||
| Jackknife | Jackknife | Jackknife | Jackknife | Jackknife | ||||
| Accuracy (%) | Accuracy (%) | Accuracy (%) | MCC | Accuracy (%) | MCC | Accuracy (%) | MCC | |
| Acrosome | 7.14 | 7.14 | 57.14 | 0.8526 | 71.43 | 0.8449 | 64.29 | 0.8659 |
| Cell membrane | 64.85 | 80.49 | 84.52 | 0.9123 | 96.67 | 0.8558 | 85.09 | 0.9121 |
| Cell wall | 12.24 | 16.33 | 91.84 | 0.8750 | 85.71 | 0.8981 | 91.84 | 0.8750 |
| Centrosome | 22.92 | 69.79 | 86.17 | 0.8650 | 92.55 | 0.6513 | 88.30 | 0.8688 |
| Chloroplast | 82.60 | 87.79 | 99.73 | 0.9943 | 99.73 | 0.9873 | 99.73 | 0.9943 |
| Cyanelle | 59.49 | 64.56 | 100.00 | 1.0000 | 98.73 | 1.0000 | 100.00 | 1.0000 |
| Cytoplasm | 64.87 | 76.72 | 45.24 | 0.9399 | 90.34 | 0.8198 | 45.70 | 0.9361 |
| Cytoskeleton | 31.65 | 27.34 | 50.36 | 0.7629 | 6.47 | 0.8318 | 49.64 | 0.7640 |
| Endoplasmic reticulum | 76.15 | 89.06 | 87.72 | 0.9529 | 84.65 | 0.9457 | 87.72 | 0.9542 |
| Endosome | 4.88 | 7.32 | 21.95 | 0.7272 | 19.51 | 0.8163 | 21.95 | 0.7497 |
| Extracell | 81.87 | 90.46 | 91.82 | 0.9812 | 88.64 | 0.9902 | 91.92 | 0.9824 |
| Golgi apparatus | 22.05 | 63.39 | 76.59 | 0.8997 | 46.83 | 0.9633 | 77.38 | 0.9131 |
| Hydrogenosome | 20.00 | 0.00 | 100.00 | 1.0000 | 70.00 | 1.0000 | 100.00 | 1.0000 |
| Lysosome | 45.61 | 31.58 | 87.72 | 0.8813 | 57.89 | 0.9851 | 87.72 | 0.8813 |
| Melanosome | 0.00 | 2.13 | 76.60 | 0.9474 | 14.89 | 1.0000 | 76.60 | 0.9474 |
| Microsome | 7.69 | 0.00 | 69.23 | 0.8579 | 15.38 | 1.0000 | 69.23 | 0.8579 |
| Mitochondrion | 70.00 | 77.05 | 78.03 | 0.9749 | 80.66 | 0.9688 | 78.20 | 0.9750 |
| Nucleus | 64.70 | 87.93 | 93.69 | 0.8865 | 50.65 | 0.9943 | 93.60 | 0.8873 |
| Peroxisome | 50.91 | 54.55 | 100.00 | 0.9650 | 74.55 | 1.0000 | 100.00 | 0.9650 |
| Spindle pole body | 33.82 | 66.18 | 95.59 | 0.9110 | 4.41 | 1.0000 | 95.59 | 0.9181 |
| Synapse | 0.00 | 38.30 | 80.85 | 0.7918 | 25.53 | 0.8399 | 80.85 | 0.7918 |
| Vacuole | 59.41 | 71.76 | 95.88 | 0.9399 | 80.59 | 0.9819 | 93.53 | 0.9606 |
| Overall accuracy | 64.17 | 79.06 | 78.01 | - | 74.70 | - | 78.17 | - |
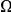
|
- | 71.27 | 75.54 | - | 72.84 | - | 75.64 | - |
