Abstract
In this paper, we introduce JGromacs, a Java API (Application Programming Interface) that facilitates the development of cross-platform data analysis applications for Molecular Dynamics (MD) simulations. The API supports parsing and writing file formats applied by GROMACS (GROningen MAchine for Chemical Simulations), one of the most widely used MD simulation packages. JGromacs builds on the strengths of object-oriented programming in Java by providing a multilevel object-oriented representation of simulation data to integrate and interconvert sequence, structure, and dynamics information. The easy-to-learn, easy-to-use, and easy-to-extend framework is intended to simplify and accelerate the implementation and development of complex data analysis algorithms. Furthermore, a basic analysis toolkit is included in the package. The programmer is also provided with simple tools (e.g., XML-based configuration) to create applications with a user interface resembling the command-line interface of GROMACS applications. Availability: JGromacs and detailed documentation is freely available from http://sbcb.bioch.ox.ac.uk/jgromacs under a GPLv3 license.
1. Introduction
Molecular dynamics (MD) simulations provide a powerful tool to study the native dynamics of biological macromolecules with atomistic resolution.1,2 Due to recent advances in hardware and software, as well as the development of enhanced sampling techniques, computer simulations can now sample biologically relevant time scales (microsecond and beyond).3 On the other hand, while simulations can better explore the conformational space of interest, the large number of conformations sampled requires increasingly sophisticated methods for analysis.4
GROMACS (GROningen MAchine for Chemical Simulations)5 is one of the four most commonly used molecular dynamics simulation suites (together with CHARMM,6 AMBER,7 and NAMD8). However, GROMACS is the only package of the four that is open-source. The GROMACS suite also includes a series of tools to process and analyze trajectories generated by simulations. Although these in-built tools cover a wide spectrum of standard analysis methods (from principal component analysis (PCA) to density calculations to clustering), one may need to develop their own analytical tools that process GROMACS trajectories. Even though it is possible to modify or extend the open source GROMACS code written in C, it would often be more convenient to build applications from scratch that operate on GROMACS data files.
The Java API (Application Programming Interface) introduced in this paper is intended to provide full freedom in developing data analysis tools that can directly process GROMACS data. The library contains native parsers for some GROMACS file formats while trajectories can be parsed via the use of gmxdump allowing simulation data to be accessed through the Java code. Data read from input files are stored in an object-oriented architecture representing different levels of structural information (from sequences to structures and trajectories). Processed data can be saved to GROMACS formats enabling integration of GROMACS and Java-based tools into a data analysis pipeline.
Our goal is to simplify the analysis of protein motions within the framework of Java, one of the most popular programming languages in academic software development and, in particular, bioinformatics. One reason for the popularity of Java is that it makes cross-platform GUI application development very easy, and GUIs are often essential to visualizing bioinformatics results. At the same time, Java is a powerful and robust object-oriented language.9 Many existing bioinformatics tools and packages were written in Java (including programming libraries such as BioJava;10 analysis and visualization tools such as StatAlign,11 Jmol,12 or Jalview;13 and complete bioinformatics analysis platforms such as Geneious14).
BioJava is a mature open-source project providing a framework for the analysis of biological data in general. It provides Java classes representing biological objects and a large collection of analytical and statistical routines covering a wide range of fields of bioinformatics. By contrast, JGromacs is designed to focus on the particular problem of processing and analyzing molecular dynamics (MD) trajectories; therefore, it is a much smaller API with more focused functionality. Packages developed for similar purposes in different programming languages include MDAnalysis15 and MMTK (Molecular Modeling Toolkit)16 designed for Python, LOOS (Lightweight Object-Oriented Structure library)17 designed for C++, and OpenStructure18 designed for Python/C++. While all frameworks mentioned offer object-oriented design, they have different support for reading and writing trajectory and coordinate file formats. From this point of view, MDAnalysis and LOOS are the most versatile, as they can import and export formats used by multiple MD suites such as Gromacs, CHARMM, AMBER, and NAMD. Unlike the other three packages, MMTK also enables setting up and running MD simulations. MDAnalysis, LOOS, and OpenStructure all offer an atom selection feature; i.e., atom groups can be selected using descriptors and boolean operators. Since JGromacs has been designed to process Gromacs trajectories, it defines atom groups via index sets used by Gromacs tools. By contrast to other packages, it also supports input/output of sequences and multiple alignments and enables the joint analysis of sequence and structural/dynamics data.
In our paper, we will first discuss the structure and main features of the JGromacs application programming interface (API). It is followed by an example presenting a simple JGromacs code and its application on a sample MD trajectory. As illustrated in the example below, complicated concepts that would normally take hours to code up from scratch can be implemented in a matter of minutes with the help of the JGromacs library.
2. Structure and Features of the API
2.1. Object-Oriented Description
The JGromacs library comprises 5 subpackages, each of which is a collection of Java classes sharing a distinct function. The core subpackage, jgromacs.data, contains 13 classes representing different levels of structural data from single atoms and amino acid residues to protein structures to complete MD trajectories. The subpackage also contains classes to handle amino acid sequences, multiple sequence alignments, atomic index sets, simulation frame index sets, and mathematical objects such as three-dimensional points, point sets, angles, matrices, and vectors. The objects defined in jgromacs.data are the basic building blocks of JGromacs applications and can be interconverted between each other in many ways.
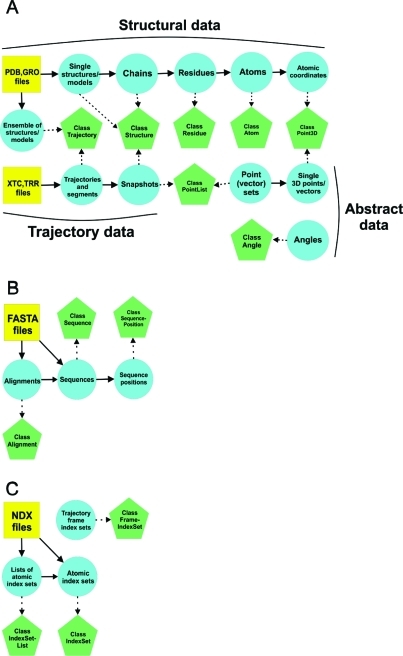
Figure 1 shows how these hierarchically related classes represent multiple levels of sequence, structure, and trajectory information. The class Structure, for example, can be used to store single structural models read from coordinate files and separate polypeptide chains. A Structure object wraps a collection of Residue objects that represent amino acid residues, water, and other molecules in the structure. On the other hand, a Residue object wraps a collection of Atom objects representing the atoms in the residue. Atomic coordinates are stored by objects of the Point3D class. JGromacs defines groups of atoms with the help of index sets, analogously to the index (.NDX) files in GROMACS.
Figure 1.
JGromacs classes and multiple levels of data represented: (A) structures and trajectories, (B) sequences and alignments, (C) atomic index sets and MD frame index sets. Blue circles depict different levels of information; green pentagons depict Java classes. Arrows between circles show hierarchical relationships, while arrows between circles and pentagons indicate mapping between data and JGromacs objects.
MD trajectories and structural (e.g., NMR) ensembles are stored in objects of the Trajectory class. Frames of a trajectory can be retrieved either as Structure or PointList objects which are used to extract atomic coordinates. The Sequence and Alignment classes are designed to represent amino acid sequences and multiple sequence alignments. Atom and residue types are defined in subpackage jgromacs.db.
The classes in jgromacs.data provide methods for retrieving and modifying the properties of data objects such as rotating and translating atoms, calculating interatomic and inter-residue distances, extracting trajectory segments, retrieving an amino acid sequence from a protein, etc. For further information on the functionalities of subpackage jgromacs.data, see the API’s documentation (also available in the SI).
2.2. Parsing GROMACS Files
The jgromacs.io subpackage provides native parsers for PDB, GRO, and NDX. XTC and TRR formats are parsed via use of the gmxdump package within GROMACS. This enables JGromacs to import structures, trajectories, and index groups to JGromacs objects. Structures and index sets can be saved back to GROMACS files with the output routines of jgromacs.io. The jgromacs.io subpackage also offers parsers and output functions for FASTA format to import and export sequences and alignments. Importing and exporting data between GROMACS files and JGromacs objects enables us to connect Java tools and GROMACS tools in an integrated data analysis pipeline. Furthermore, the subpackage jgromacs.io provides an option to execute any GROMACS commands from within the Java code and automatically import the output files as JGromacs objects.
2.3. In-Built Analysis Toolkit
The subpackage jgromacs.analysis offers a collection of analytical routines covering various areas from calculating dihedral angles to extracting contact matrices to weighted superposition of structures. Making use of the toolkit, one can for example retrieve the mean distance matrix or covariance matrix of a trajectory, calculate the root-mean-square inner product (RMSIP) between conformational subspaces, look at the cumulative variance profiles in PCA, extract time series of interatomic distances or dihedral angles, find the simulation snapshot where two atoms are in closest proximity, use Gaussian network models, and many more. These analysis functions operate on the objects defined in subpackage jgromacs.data. The toolkit can easily be extended with additional routines that fit into this framework.
2.4. User Interface Support
Finally, subpackage jgromacs.ui provides a simple way to add a user-friendly interface to JGromacs applications. The user interface (UI) can easily be set up with an XML configuration file. It supports help messages, log files, and command line argument parsing and in many aspects resembles the UI of GROMACS tools.
3. An Example: Dynamical Networks
An example is presented below to illustrate how JGromacs simplifies the implementation of complex ideas such as the concept of dynamical networks.19
3.1. Dynamical Networks
The definition of dynamical networks was introduced by Sethi et al. (2009, PNAS) to study allosteric signaling in tRNA:protein complexes. Their idea was to represent a tRNA:protein complex as a weighted graph in which each amino acid residue and nucleotide of the complex is represented by a single node. Two nodes are connected in the network if the monomers are in contact; i.e., their closest heavy atoms are within 4.5 Å of each other for at least 75% of the MD simulation frames. An edge between nodes i and j is weighted by the absolute value of the Cij correlation between the two monomers calculated over the course of the MD simulation. The weight of a link estimates the probability of information transfer between the two residues. The “length” of a link was defined as −log|Cij|. Adding information about dynamics, these networks give a more realistic picture about the system than the unweighted protein structure networks (PSN) constructed on the basis of the contact pattern of a single structure. Sethi et al. used network analysis concepts (i.e., shortest path, betweenness centrality, suboptimal path, and community analysis) to identify nodes and paths in the network crucial for intramolecular signal transduction, highlighting possible allosteric communication pathways within the complex.
3.2. Implementation in JGromacs
The following
13-line JGromacs code calculates the weight matrix of the dynamical
network of a protein from a GROMACS MD trajectory:
As a first step, the example code imports structure and trajectory data from GRO and XTC files. It then determines the frequency-based contact matrix using a 0.45 nm distance cutoff and a 0.75 contact probability cutoff. After extracting the trajectory of α carbon atoms, it calculates their correlation matrix. Finally, the contact and correlation matrices are combined into the output matrix W that defines the connectivity and weights of the dynamical network. The weight matrix W is the input of further analysis.
3.3. Application to Example Data
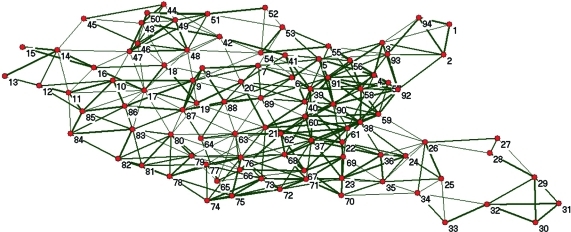
Figure 2 shows the dynamical network of the N-terminal PDZ domain of InaD (Inactivation no afterpotential D) protein from Drosophila based on a 20 ns molecular dynamics simulation. The topology and the weights of the graph were calculated with the short JGromacs code above. Figure 2 was created using the network analysis and visualization software Pajek.20 Starting from scratch, generating this network from an MD trajectory file would be time-consuming, but JGromacs significantly reduces programming time. Further examples and a step-by-step Quick Start Guide are downloadable from the project Web site.
Figure 2.
Dynamical network created for the PDZ1 domain of InaD protein from Drosophila based on a 20 ns MD simulation. Nodes represent residues; edge widths are proportional to link weights.
4. Conclusions
As computer simulations are becoming more and more effective in sampling the conformational dynamics of biological macromolecules, the storage, management, and analysis of the generated data present an ever-increasing challenge. There have been not only efforts to address the storage issues21 but also an additional analysis suite as found in the BioSimGrid platform.22 The analysis toolkit of BioSimGrid is an extensive collection of standard analysis routines (e.g., root-mean-square deviations, volume and average structure, interatomic distance,s and surface area) facilitating cross-comparison of the deposited trajectories. On the other hand, molecular dynamics software packages such as GROMACS and CHARMM have their own in-built analysis tools providing the significant advantage of performing simulations and analysis within the same framework. However, in addition to making use of the standard analysis routines implemented in these platforms, one may also need a flexible framework for developing their own novel tools for analyzing MD data.
JGromacs is a lightweight Java library supporting simple and fast development of analytical tools for data sets produced with the commonly used MD software GROMACS. The objective of our project is to create a framework for implementing increasingly complex analytical routines that can be used through simple user interfaces. Since in research the goal is not always to develop ready-made applications but to experiment with new ideas as quickly as possible, simplicity of the package was of utmost importance.
While JGromacs also contains a standard analysis toolkit, its main advantage is that it provides an object-oriented framework for novel tool development. The programmers can easily build up their own algorithms and applications based on the basic JGromacs classes and analytical routines already implemented in the package. Furthermore, the library provides options for integrating Java and GROMACS analysis tools.
A detailed documentation (including Quick Start Guide, examples and description of all subpackages, classes, and methods), Javadoc (HTML) documentation, a comprehensive JUnit test suite, a library of executable example codes, and an example data set are available on the project Web site: http://sbcb.bioch.ox.ac.uk/jgromacs/.
Acknowledgments
P.C.B. thanks the Wellcome Trust for support. M.M. thanks the BBSRC via the Systems Biology Doctoral Training Centre.
Supporting Information Available
Complete documentation and data sets are given as Supporting Information as outlined below:
-
1.
Complete documentation of JGromacs v1.0. API (PDF file): jgromacs_v1_doc.pdf
-
2.
JavaDoc documentation of JGromacs v1.0. API (gzipped tar file): jgromacs_v1_javadoc.zip
-
3.
Library of example codes (gzipped tar file): jgromacs_v1_examples.zip
-
4.
Test suite: jgromacs_v1_test.zip
This information is available free of charge via the Internet at http://pubs.acs.org/.
Supplementary Material
References
- Karplus M.; McCammon J. A. Molecular Dynamics Simulations of Biomolecules. Nat. Struct. Biol. 2002, 9, 646–652. [DOI] [PubMed] [Google Scholar]
- Cascella M.; Dal Peraro M. Challenges and Perspectives in Biomolecular Simulations: From the Atomistic Picture to Multiscale Modeling. Chim. Int. J. Chem. 2009, 63, 14–18. [Google Scholar]
- Zwier M. C.; Chong L. T. Reaching Biological Timescales with All-Atom Molecular Dynamics Simulations. Curr. Opin. Pharmacol. 2010, 10, 745–752. [DOI] [PubMed] [Google Scholar]
- Salsbury F. R. Jr. Molecular Dynamics Simulations of Protein Dynamics and Their Relevance to Drug Discovery. Curr. Opin. Pharmacol. 2010, 10, 738–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B.; Kutzner C.; van der Spoel D.; Lindahl E. Gromacs 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simuation. J. Chem. Theory Comput. 2008, 4, 435–447. [DOI] [PubMed] [Google Scholar]
- Brooks B. R.; Bruccoleri R. E.; Olafson B. D.; States D. J.; Swaminathan S.; Karplus M. Charmm: A Program for Macromolecular Energy, Minimisation, and Dynamics Calculations. J. Comput. Chem. 1983, 4, 187–217. [Google Scholar]
- Pearlman D. A.; Case D. A.; Caldwell J. W.; Ross W. S.; Cheatham T. E.; Debolt S.; Ferguson D.; Seibel G.; Kollman P. Amber, a Package of Computer-Programs for Applying Molecular Mechanics, Normal-Mode Analysis, Molecular-Dynamics and Free-Energy Calculations to Simulate the Structural and Energetic Properties of Molecules. Comput. Phys. Commun. 1995, 91, 1–41. [Google Scholar]
- Phillips J. C.; Braun R.; Wang W.; Gumbart J.; Tajkhorshid E.; Villa E.; Chipot C.; Skeel R. D.; Kale L.; Schulten K. Scalable Molecular Dynamics with Namd. J. Comput. Chem. 2005, 26, 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosling J.; Mcgilton H.. The Java Language Environment: A White Paper; Sun Microsystems: Santa Clara, CA, 1996. [Google Scholar]
- Holland R. C.; Down T. A.; Pocock M.; Prlic A.; Huen D.; James K.; Foisy S.; Drager A.; Yates A.; Heuer M.; Schreiber M. J. Biojava: An Open-Source Framework for Bioinformatics. Bioinformatics 2008, 24, 2096–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak A.; Miklos I.; Lyngso R.; Hein J. Statalign: An Extendable Software Package for Joint Bayesian Estimation of Alignments and Evolutionary Trees. Bioinformatics 2008, 24, 2403–2404. [DOI] [PubMed] [Google Scholar]
- Hanson R. M. Jmol - a Paradigm Shift in Crystallographic Visualization. J. Appl. Crystallogr. 2010, 43, 1250–1260. [Google Scholar]
- Waterhouse A. M.; Procter J. B.; Martin D. M.; Clamp M.; Barton G. J. Jalview Version 2--a Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J.; Ashton B.; Buxton S.; Cheung M; Cooper A.; Heled J.; Kearse M.; Sturrock S.; Thierer T.; Wilson A. Geneious V5.1. http://www.geneious.com (accessed Sept 21, 2011).
- Michaud-Agrawal N.; Denning E. J.; Woolf T.; Beckstein O. Mdanalysis: A Toolkit for the Analysis of Molecular Dynamics Simulations. J. Comput. Chem. 2011, 32, 2319–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsen K. The Molecular Modelling Toolkit: A New Approach to Molecular Simulations. J. Comput. Chem. 2000, 21, 79–85. [Google Scholar]
- Romo T. D.; Grossfield A.. In Loos: An Extensible Platform for the Structural Analysis of Simulations. IEEE Eng. Med. Biol. Soc. 2009, 2332–2335. [DOI] [PubMed] [Google Scholar]
- Biasini M.; Mariani V.; Haas J.; Scheuber S.; Schenk A. D.; Schwede T.; Philippsen A. Openstructure: A Flexible Software Framework for Computational Structural Biology. Bioinformatics 2010, 26, 2626–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi A.; Eargle J.; Black A. A.; Luthey-Schulten Z. Dynamical Networks in Trna:Protein Complexes. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 6620–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batagelj V.; Mrvar A. Pajek - Program for Large Network Analysis. Connections 1998, 21, 47–57. [Google Scholar]
- Vohra S.; Hall B. A.; Holdbrook D. A.; Khalid S.; Biggin P. C., Bookshelf: A Simple Curation System for the Storage of Biomolecular Simulation Data. Database. [online] 2010, 2010, baq033. http://database.oxfordjournals.org/content/2010/baq033.abstract (accessed Sept 21, 2011). [DOI] [PMC free article] [PubMed]
- Tai K.; Murdock S.; Wu B.; Ng M. H.; Johnston S.; Fangohr H.; Cox S. J.; Jeffreys P.; Essex J. W.; Sansom M. S. P. Biosimgrid: Towards a Worldwide Repository for Biomolecular Simulations. Org. Biomol. Chem. 2004, 2, 3219–3221. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.