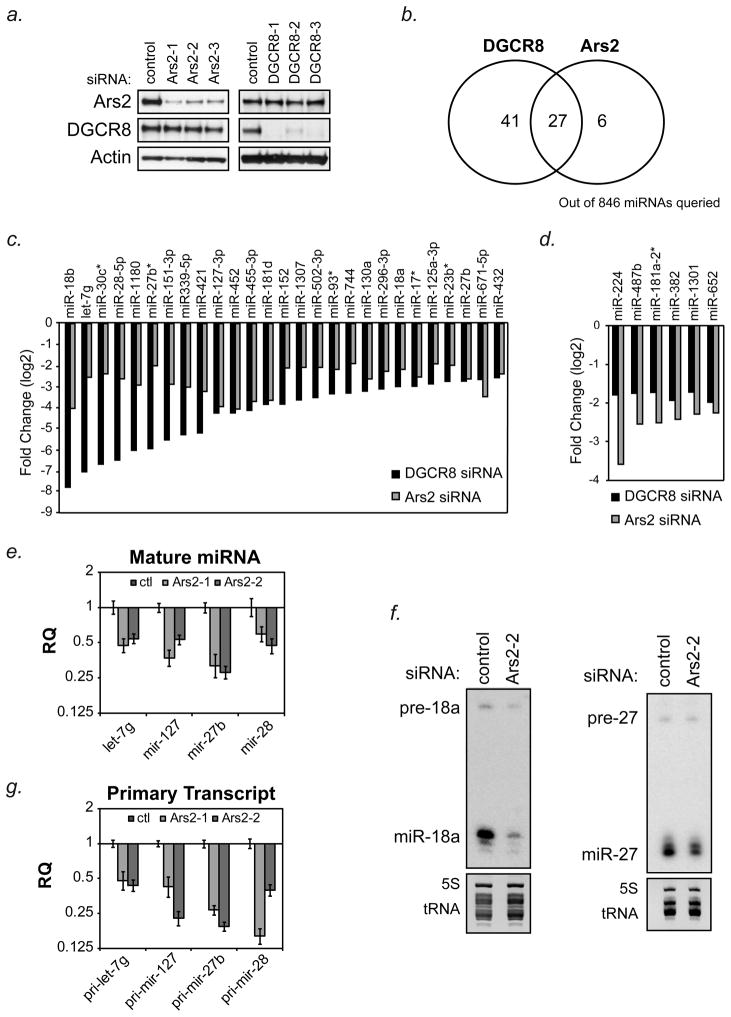
Figure 1. Ars2 regulates a subset of microRNAs.
(A) HeLa cells were transfected with control siRNA or 3 siRNAs targeted to either Ars2 (Ars2-1, Ars2-2, Ars2-3) or DGCR8 (DGCR8-1, DGCR8-2, DGCR8-3). Protein was harvested three days following transfection to confirm specific depletion of Ars2 or DGCR8 by Western blot.
(B) HeLa cells were transfected with the siRNAs as in (A) and RNA was isolated three days later and analyzed separately on Affymetrix GeneChip® miRNA Arrays. The number of microRNAs decreased at least 2-fold (log2) following transfection of all three siRNAs per gene are depicted by Venn Diagram.
(C) Bar graphs showing 27 microRNAs determined by microarray to decrease 2-fold (log2) or more following depletion of DGCR8 or Ars2. Bars represent the average of the three siRNAs targeting DGCR8 or Ars2 shown in (A).
(D) Bar graphs showing 6 microRNAs determined by microarray to decrease 2-fold (log2 ) or more following depletion of Ars2 but not DGCR8. Bars represent the average of the three siRNAs targeting DGCR8 or Ars2 shown in (A).
(E) HeLa cells were transfected with two siRNAs targeted to Ars2 (Ars2-1, Ars2-2) or a control siRNA (ctl). Three days later RNA was isolated and TaqMan® microRNA assays were used to detect the mature microRNAs indicated. Bars represent relative quantification using the ΔΔCt method +/− 95% confidence interval of three replicates. U6 snRNA was used as an endogenous control.
(F) HeLa cells were transfected with siRNA to Ars2 (Ars2-2) or a control siRNA (ctl) and three days later RNA was isolated. Precursor and mature microRNAs indicated were detected by Northern blot. Ethidium bromide staining of 5S rRNA and tRNAs are shown as loading controls.
(G) RNA from (E) was reverse transcribed and TaqMan®-based qPCR was performed for the indicated primary microRNA transcripts. Bars represent relative quantification using the ΔΔCt method +/− 95% confidence interval of three replicates. U6 snRNA was used as an endogenous control.