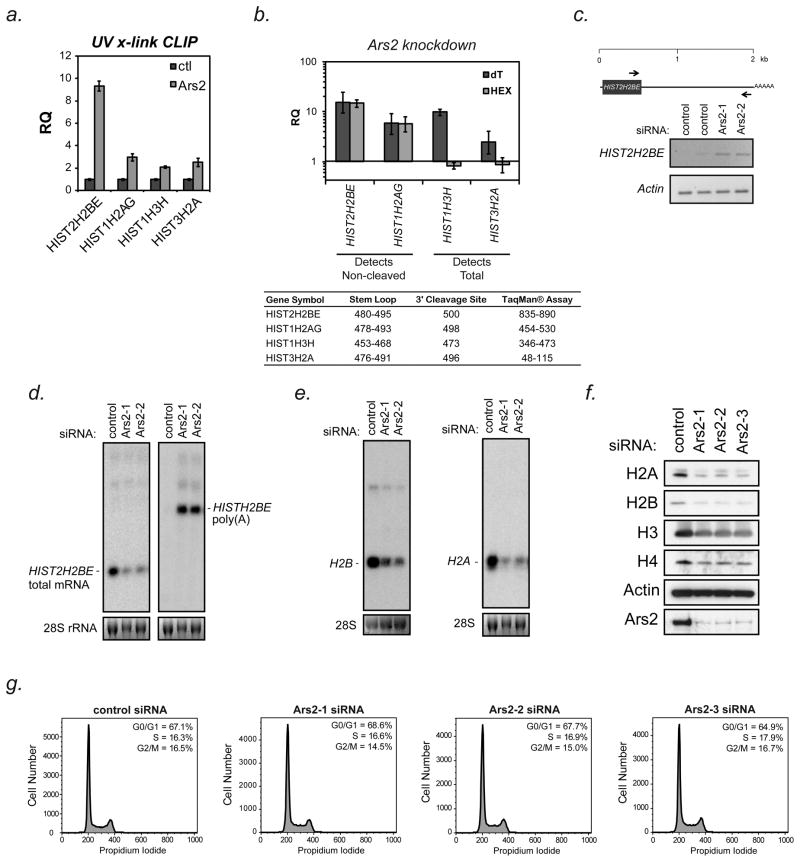
Figure 3. Ars2 binds to and regulates the expression and processing of replication-dependent histone transcripts.
(A) To determine if Ars2 could directly associate with replication-dependent histone mRNAs, ultraviolet light crosslinking followed by immunoprecipitation (UV-CLIP) with a monoclonal antibody to Ars2 (9A12) or control antibody (SP/0) was performed. RNA isolated from the precipitates was reverse transcribed with random hexamer primers and used for qPCR with TaqMan® primer/probe sets to the indicated genes. Bars represent relative quantification using the ΔΔCt method normalized to control antibody CLIP +/− 95% confidence interval of three replicates. Non-specifically bound 18S rRNA was used as an endogenous control.
(B) HeLa cells were transfected with siRNA to Ars2 (Ars2-3) or a control siRNA and 3 days later RNA was isolated. cDNA synthesis was performed using either oligo(dT) or random hexamer primers. Transcript levels were measured by TaqMan®-based qPCR using the assays outlined in the inset table. Because the TaqMan® assays used for HIST2H2BE and HIST2H2AG amplified a region downstream of their 3′ cleavage sites they only detected non-cleaved transcripts (i.e. those that extend beyond the normal 3′ cleavage sites), whereas the assays for HIST1H3H and HIST3H2A were upstream of their 3′ cleavage sites and could therefore differentiate between polyadenylated transcripts (dT) and total mRNA expression (HEX). Bars represent relative quantification using the ΔΔCt method normalized to control siRNA transfection +/− 95% confidence interval of three replicates. Human ACTB was used as an endogenous control.
(C) Top - Schematic of the human HIST2H2BE gene with scale in kilobases (kb). The arrows indicate the position of primers used for PCR analysis. Bottom - HeLa cells were transfected with siRNAs to Ars2 (Ars2-1, Ars2-2) or control siRNAs and three days later RNA was isolated. PCR was performed following oligo(dT) reverse-transcription using the primers indicated in the schematic. Actin was amplified as a control.
(D) HeLa cells were transfected with control or Ars2 siRNAs (Ars2-1, Ars2-2), RNA was isolated after 3 days and Northern blotting was performed for the HIST2H2BE transcript. Left - Total HIST2H2BE mRNA was detected with a 34 nucleotide probe designed to hybridize selectively to the HIST2H2BE 3′UTR upstream of the stem-loop. Right - To detect HIST2H2BE transcripts that were not properly cleaved, the membrane was stripped and re-probed with a probe generated by nick translation of a cDNA of the HIST2H2BE 3′UTR.
(E) Additional Northern blots were performed on RNA isolated from HeLa cells transfected with Ars2 siRNAs (Ars2-1, Ars2-2) or control siRNA. Blots were probed with oligonucleotide probes within the open reading frames of histone H2B (left) or H2A (right) genes and therefore detect multiple conserved transcripts.
(F) HeLa cells were transfected with control or three different Ars2 siRNAs (Ars2-1, Ars2-2, Ars2-3) and proteins were harvested after three days. Western blotting was performed for the four core histone proteins. Knockdown was confirmed by probing for Ars2 and Actin was used as a loading control.
(G) Cell cycle analysis was performed on HeLa cells three days after transfection with control or three independent siRNAs targeting Ars2 (Ars2-1, Ars2-2, Ars2-3). Quantification of cells in G0/G1 (1N), G2 (2N) and S-phase (intermediate) of the cell cycle is shown in the upper right corner of each histogram. In agreement with these data the amount of DNA per cell remained unaltered following Ars2 knockdown (43.3 pg/cell in Ars2 siRNA vs. 42 pg/cell in control).