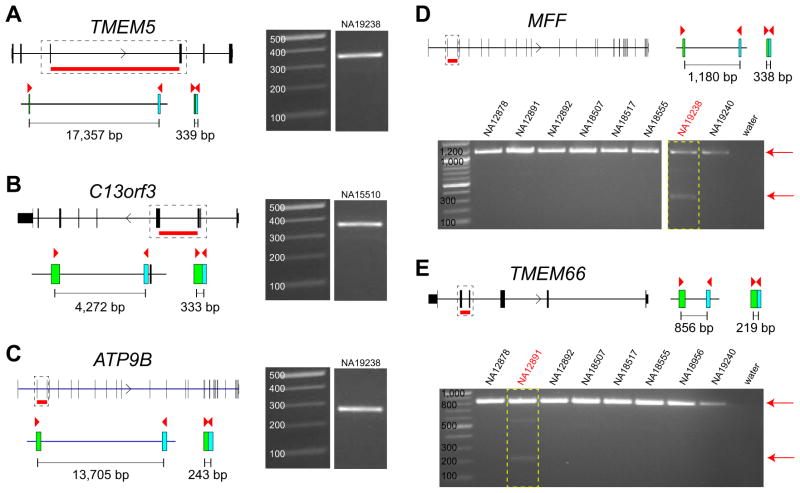
Figure 2. Validation of processed pseudogenes.
Gene models and predicted intron deletions of the processed pseudogenes are shown. Primers (red triangles) are designed in the coding region of the genes and the expected product size for the processed pseudogenes are shown for (A) TMEM5, (B) C13orf3, (C) ATP9B, (D) MFF, and (E) TMEM66. Gel images show the size of the amplified product. We were able to detect the processed version of these genes in our PCR experiments. In D-E we genotyped the processed pseudogenes MFF and TMEM66 within eight HapMap samples and show that each is amplified only in the predicted sample [boxed in red: NA19238 (MFF) and NA12891 (TMEM66)]. All PCRs amplify the normal gene (signal on the top) with only one sample each amplifying the processed gene.