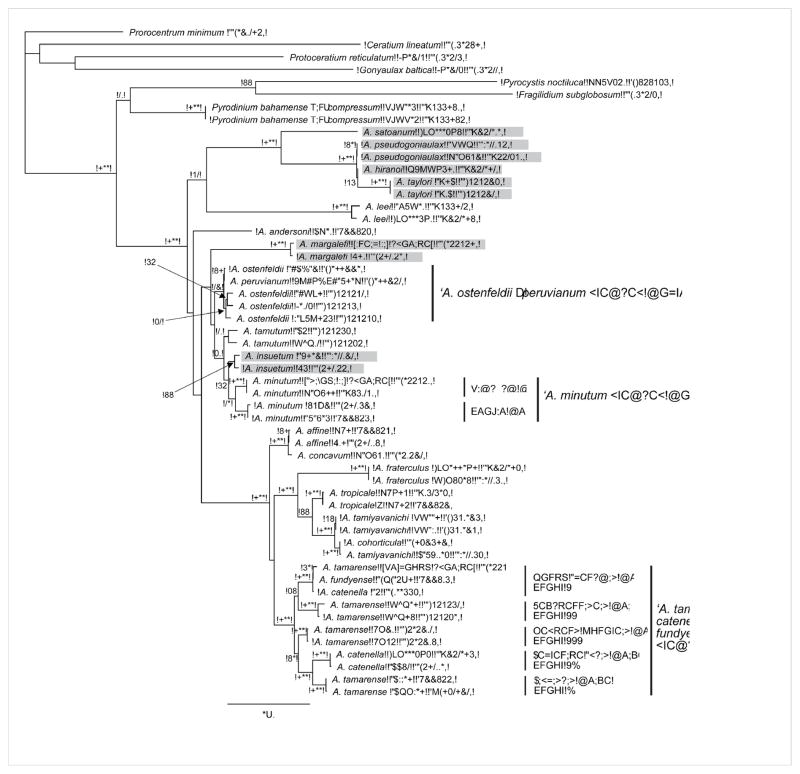
Figure 1.
Phylogenetic tree inferred by maximum likelihood analysis of partial LSU rDNA (D1–D2 domains) of 21 nominal species of Alexandrium. Analysis includes a subset of taxa included in the maximum likelihood phylogenetic analysis of 28S rDNA by Touzet et al. (2008a). This analysis was supplemented by additional sequences for some species (or ribotypes of species complexes) from previous phylogenetic studies: A. pseudogonyaulax (MacKenzie et al., 2004), A. tropicale and A. minutum ‘Pacific clade’ (Lilly et al., 2005), A. ostenfeldii (Kremp et al., 2009), A. tamutum (Montresor et al., 2004), A. fraterculus and A. taylori (John et al., 2003b), and A. tropicale and A. tamiyavanichi (Menezes et al., 2010). In addition, Pyrodinium bahamense sequences used in the analyses by Leaw et al. (2005) were included, as well as those of other gonyaulacoid dinoflagellates, to demonstrate monophyly of the genus Alexandrium. Prorocentrum minimum was set as the outgroup. Sequences were aligned with MAFFT v6.814b (Katoh and Kuma, 2002) in Geneious 5.4.4 and the TrN+G model of base substitution was determined according to the Akaike Information Criterion and the Bayesian Information Criterion as the optimal model with jModeltest (Posada, 2008). Maximum likelihood analyses were carried out with PhyML (Guindon and Gascuel 2003) in Geneious 5.4.4 with the following constraining parameters: base frequency (A= 0.26832, C= 0.15771, G= 0.25629, T= 0.31768), Transition/transversion ratio for purines: 2.267, Transition/transversion ratio for pyrimidines: 4.725, gamma distribution shape parameter (G= 0.755). Branch frequencies from 100 bootstrap replicates are given in percent at the respective nodes if >50%. The two subgenera Alexandrium and Gessnerium (light gray shaded) do not form reciprocal monophyletic clades. Species complexes, such as the A. tamarense species complex, contain non-reciprocal monophyletic clades according to morphologically determined taxa, which rather resemble evolutionary units with distinct biogeographical distributions and varying degrees of morphological plasticity.
* Isolate was originally misidentified as A. tropicale (Lilly et al., 2007)