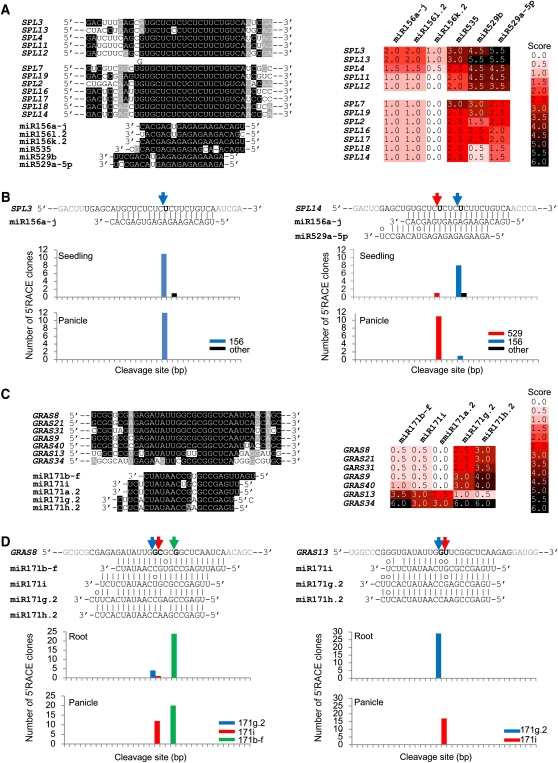
Figure 9.
Differential Selection of Target Genes by Differentially Expressed miRNAs.
(A) Sequence alignment of miR156 family members and their target genes. The table next to the target genes represents target score for each member of the miR156 family.
(B) Validation of target cleavage by RLM 5′-RACE. Predicted cleavage sites are indicated by a bold nucleotide at position 10 relative to the 5′ end of the miRNA. The number of cloned RACE products sequenced is shown in the histogram.
(C) Sequence alignment of the miR171 family and their target genes.
(D) Validation of target cleavage by the miR171 family. The cleavage of GRAS13 shown formiR171g.2 could be equally the result of miR171 h.2. Names of target genes were from a previous publication (Tian et al., 2004), and their Michigan State University Rice Genome Annotation Project locus names are shown in Supplemental Table 8 online.