PMD1 was identified as a novel plant-specific coiled-coil protein that is tail anchored to the membrane of peroxisomes and mitochondria, capable of inducing the proliferation of both organelles. Its homolog, PMD2, exclusively targets to mitochondria and is involved in the morphogenesis of mitochondria.
Abstract
Peroxisomes and mitochondria are multifunctional eukaryotic organelles that are not only interconnected metabolically but also share proteins in division. Two evolutionarily conserved division factors, dynamin-related protein (DRP) and its organelle anchor FISSION1 (FIS1), mediate the fission of both peroxisomes and mitochondria. Here, we identified and characterized a plant-specific protein shared by these two types of organelles. The Arabidopsis thaliana PEROXISOMAL and MITOCHONDRIAL DIVISION FACTOR1 (PMD1) is a coiled-coil protein tethered to the membranes of peroxisomes and mitochondria by its C terminus. Null mutants of PMD1 contain enlarged peroxisomes and elongated mitochondria, and plants overexpressing PMD1 have an increased number of these organelles that are smaller in size and often aggregated. PMD1 lacks physical interaction with the known division proteins DRP3 and FIS1; it is also not required for DRP3’s organelle targeting. Affinity purifications pulled down PMD1’s homolog, PMD2, which exclusively targets to mitochondria and plays a specific role in mitochondrial morphogenesis. PMD1 and PMD2 can form homo- and heterocomplexes. Organelle targeting signals reside in the C termini of these proteins. Our results suggest that PMD1 facilitates peroxisomal and mitochondrial proliferation in a FIS1/DRP3-independent manner and that the homologous proteins PMD1 and PMD2 perform nonredundant functions in organelle morphogenesis.
INTRODUCTION
In eukaryotic cells, organelles are delimited by their own lipid bilayers, providing membrane-bound compartments for specific biochemical reactions to occur. Peroxisomes and mitochondria are ubiquitous and multifunctional organelles with essential roles in development. Surrounded by a single membrane, peroxisomes house a variety of metabolic processes, such as fatty acid β-oxidation, scavenging of reactive oxygen species and peroxides, ether phospholipid biosynthesis, and fatty acid α-oxidation in mammals and photorespiration and the glyoxylate cycle in plants (Wanders and Waterham, 2006; Kaur et al., 2009). Mitochondria are enclosed by a double membrane and serve as the powerhouse of the cell by performing functions such as respiration, ATP synthesis, and tricarboxylic acid cycle (Millar et al., 2008). Although each type of organelle carries a unique set of biochemical functions, a number of intracellular metabolic pathways are known to be completed coordinately by multiple organelles, including peroxisomes and mitochondria. In plants, for example, the recycling of phosphoglycolate during photorespiration is executed by the sequential action of chloroplasts, peroxisomes, and mitochondria (Peterhansel et al., 2010). The conversion of fatty acids to Suc during oilseed establishment involves the cooperative participation of lipid bodies, peroxisomes, mitochondria, and the cytosol (Baker et al., 2006; Penfield et al., 2006).
In light of these highly coupled functions, it is not that surprising that peroxisomes and mitochondria also share division factors (Delille et al., 2009; Kaur and Hu, 2009). The peroxisome is believed to be an endoplasmic reticulum (ER)–derived member of the endomembrane system and can form out of the ER in cells in which peroxisomes are lost (Hoepfner et al., 2005; Gabaldón et al., 2006; Schlüter et al., 2006; Titorenko and Mullen, 2006). Peroxisomes can also proliferate from preexisting peroxisomes through growth and division (Purdue and Lazarow, 2001; Fagarasanu et al., 2007; Kaur and Hu, 2009). Mitochondria, like chloroplasts, are descendents of ancient endosymbionts with bacterial origins and thus divide exclusively by binary fission from preexisting organelles (Osteryoung and Nunnari, 2003). Despite having distinct evolutionary histories and ultrastructures, peroxisomes and mitochondria share at least two proteins in the fission process across animal, fungal, and plant kingdoms (Fagarasanu et al., 2007; Kaur and Hu, 2009). Dynamin-related proteins (DRPs) are key factors in peroxisomal and mitochondrial division, where these large and self-assembling GTPases form a spiral-like structure around the membranous structures to mediate membrane fission through GTP hydrolysis (Praefcke and McMahon, 2004; Kaur and Hu, 2009). Through forward genetic screens followed by homology-based searches, two Arabidopsis thaliana DRP homologs, DRP3A and DRP3B, have been found to mediate the division of peroxisomes and mitochondria, with DRP3A playing a predominant role (Arimura and Tsutsumi, 2002; Arimura et al., 2004; Aung and Hu, 2009; Fujimoto et al., 2009; Zhang and Hu, 2009). DRP5B, a DRP distantly related to DRP3, was found to be localized to peroxisomes and chloroplasts and mediate the division of these two organelles, which are also linked through a number of metabolic pathways (Gao et al., 2003; Zhang and Hu, 2010). Since most eukaryotic DRPs lack a putative lipid binding domain (Pleckstrin homology domain) or transmembrane domain (TMD), they are often found in the cytosol and only recruited to the division sites by interacting directly or indirectly with a membrane-bound receptor named FISSION1 (FIS1) (reviewed in Kaur and Hu, 2009). FIS1 is tethered to the membranes by its C terminus, exposing its N-terminal tetratricopeptide repeat domain to the cytosol (Mozdy et al., 2000; Koch et al., 2003; Koch et al., 2005; Kobayashi et al., 2007). Arabidopsis contains two homologs of FIS1, FIS1A (BIGYIN) and FIS1B. Protein localization and reverse genetic analyses confirmed the role of the Arabidopsis FIS1A and FIS1B in peroxisomal and mitochondrial division, although their role in recruiting DRP3 proteins to the division sites has not been proven yet (Scott et al., 2006; Lingard et al., 2008; Zhang and Hu, 2008, 2009).
In addition to FIS1 and DRP, the yeast peroxisomal and mitochondrial division complex also contains adaptor proteins, Mdv1p and Caf4p, which are WD-40 proteins that interact with both DRP and FIS1 to target DRP to the fission sites (Tieu and Nunnari, 2000; Tieu et al., 2002; Motley et al., 2008; Nagotu et al., 2008). Although homologs of Mdv1p/Caf4p have not been identified in metazoans and plants, Mitochondrial fission factor (Mff) is a metazoan-specific and tail-anchored coiled-coil (CC) protein that is involved in the division of peroxisomes and mitochondria (Gandre-Babbe and van der Bliek, 2008). It interacts directly with Drp1 and recruits it to the mitochondrial fission sites in a Fis1-independent manner (Otera et al., 2010). In Arabidopsis, ELONGATED MITOCHONDRIA1 (ELM1) is a plant-specific protein that mediates mitochondrial division by recruiting DRP3A to the mitochondrial division sites; it does not play a role in peroxisomal division. ELM1 lacks putative TMDs and does not interact with FIS1; thus, the mechanism by which it recruits DRP3A to the mitochondrial membrane remains to be elucidated (Arimura et al., 2008). In summary, whereas the core peroxisomal and mitochondrial division factors DRP and FIS1 are conserved in eukaryotes, lineage- and organelle-specific components of the division machineries also exist.
Efforts to uncover components of the plant peroxisome division apparatus, such as forward genetic screens in Arabidopsis plants expressing a fluorescent protein tagged by the C-terminal Peroxisome Targeting Signal Type1 (PTS1), repeatedly identified alleles of DRP3A (Mano et al., 2004; Aung and Hu, 2009; Zhang and Hu, 2009). To identify additional components, we employed an in silico strategy by searching the Arabidopsis genome for uncharacterized proteins with putative TMDs in addition to a protein–protein interaction domain that is also found in previously identified organelle division proteins. We initially focused on the CC domain because several CC proteins have been shown to function in organelle division in diverse species. A CC domain consists of heptad repeats, each of which contains hydrophobic residues in the first and fourth positions and charged/polar residues in the fifth and seventh positions. CC proteins are known to homo- or heterodimerize and are involved in diverse cellular functions (Rose et al., 2004, 2005). In addition to the aforementioned mammalian mitochondrial and peroxisome division factor Mff, which is a C-terminal tail-anchored (C-TA) protein with a cytosolic CC domain (Gandre-Babbe and van der Bliek, 2008; Otera et al., 2010), Arabidopsis homologous proteins PLASTID DIVISION1 (PDV1) and PDV2 also contain CC domains. The PDV proteins are anchored to the outer envelope membrane of plastids via the TMD and use the N-terminal cytosolic CC domain to recruit the large GTPase DRP5B (ARC5) to the division site (Miyagishima et al., 2006; Yang et al., 2008).
Here, we report the identification of a CC protein, PEROXISOMAL and MITOCHONDRIAL DIVISION FACTOR1 (PMD1), from the ARABI-COIL database (http://136.227.60.226/arabidopsis/main.html; Rose et al., 2004). PMD1 is an integral membrane protein localized to both peroxisomes and mitochondria. Genetic analyses of the pmd1 mutants revealed its role in the morphogenesis and proliferation of peroxisomes and mitochondria. We also identified a PMD1 homolog, PMD2, as a PMD1-interacting protein that functions exclusively in mitochondria. The C termini of PMD1 and PMD2 constitute organelle targeting signals. Our results suggest that the plant-specific protein PMD1 contributes to peroxisomal and mitochondrial proliferation in a pathway that is independent from the previously defined pathway controlled by the FIS1-DRP3 complex. Furthermore, the homologous proteins PMD1 and PMD2 perform nonredundant functions in organelle morphogenesis.
RESULTS
PMD1 Is Dual Localized to Peroxisomes and Mitochondria, Whereas Its Homolog, PMD2, Exclusively Targets to Mitochondria
To identify additional proteins involved in peroxisome division/proliferation, we used two criteria to search the ARABI-COIL database available at http://136.227.60.226/arabidopsis/main.html (Rose et al., 2004). First, the protein has to have at least one putative TMD, as predicted by the plant membrane protein database (Aramemnon; http://aramemnon.uni-koeln.de/). Second, since it was estimated that ~10% of the total proteins of an organism contain CC motifs (Liu and Rost, 2001), we first focused on proteins that contain long CC domains (i.e., those that cover >50% of the entire protein sequence). As a result, seven proteins were retrieved. Four of them had been experimentally verified by previous studies to target to the cytosol, nucleus, plasma membrane, or plastid, two were predicted to be mitochondrial, and one was predicted to localize to multiple nonperoxisomal compartments (see Supplemental Table 1 online). We made yellow fluorescent protein (YFP) fusions of these proteins and coexpressed them transiently with a peroxisomal marker in tobacco (Nicotiana tabacum). Fluorescent microscopy of the infiltrated tobacco leaf epidermal cells was performed to identify those candidate proteins that are associated with peroxisomes. Only one protein, which is encoded by At3g58840, was found to colocalize with peroxisomes. We named this protein PMD1 because it was later found to localize to both peroxisomes and mitochondria (see below).
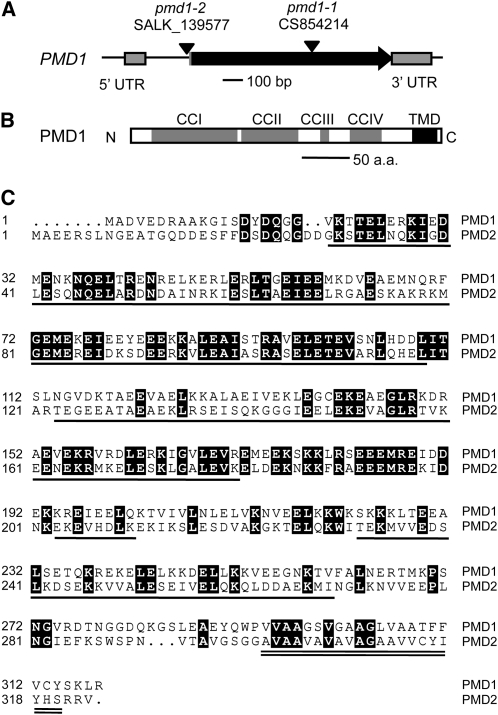
The PMD1 gene has two exons (Figure 1A). Its protein product contains four putative CC domains at the N terminus and a single segment of TMD near the C-terminal end (Figure 1B). Therefore, PMD1 qualifies as a C-TA protein, as defined previously for proteins with a single membrane-spanning domain at or near the C terminus (Abell and Mullen, 2011). Arabidopsis has a homolog of PMD1, which is encoded by At1g06530; this homolog was later named PMD2. PMD2 also has two exons and was among the seven candidate proteins identified from our in silico searches (see Supplemental Table 1 online). PMD1 and PMD2 share an overall 35% amino acid identity and similar domain structures along the length of the proteins (Figure 1C). Using protein sequence similarity-based BLAST searches, we were able to identify homologous sequences from other plant species but did not find obvious homologs of PMD1 and PMD2 in nonplant genomes.
Figure 1.
Structure and Sequence Analysis of PMD1.
(A) Genomic structure of PMD1. The T-DNA insertion sites in pmd1-1 and pmd1-2 are indicated. UTR, untranslated region.
(B) Putative protein structure of PMD1.
(C) Sequence alignment of PMD1 and PMD2. CC domains are underlined, and the TMD is double underlined. Identical sequences are shaded in black. All domain assignments are based on analysis of PMD1.
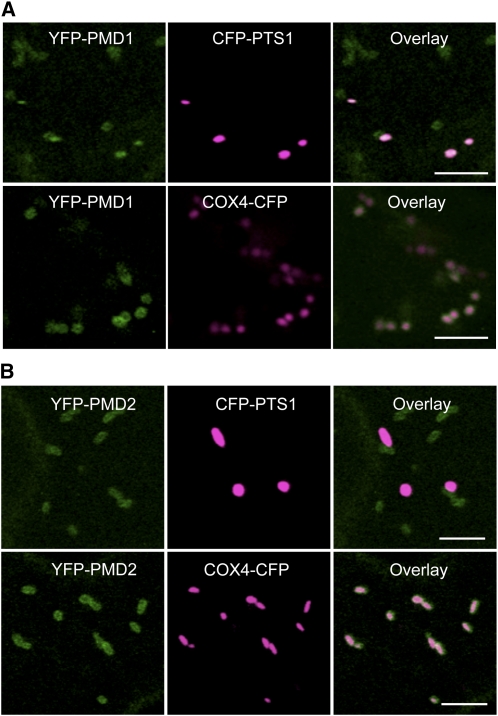
To confirm the subcellular localization of PMD1, we expressed the YFP-PMD1 fusion gene under the control of the 995-bp PMD1 native promoter in Arabidopsis plants containing a peroxisomal marker (cyan fluorescent protein [CFP]-PTS1) (Fan et al., 2005) or a mitochondrial marker (Saccharomyces cerevisiae COX4-CFP) (Nelson et al., 2007). Subcellular localization of YFP-PMD1 was examined using confocal laser scanning microscopy in T2 plants. YFP-PMD1 colocalized with CFP-PTS1 and also formed a ring-like structure on the surface of COX4-CFP–tagged mitochondria (Figure 2A). In agreement with the view that most C-TA proteins use the TMD-containing C-terminal region for targeting (Borgese et al., 2007; Abell and Mullen, 2011), the PMD1-YFP fusion protein, in which YFP is located at the C terminus of PMD1, was found only in the cytosol, possibly due to masking of the C terminus by YFP, which prevented its organelle targeting (see Supplemental Figure 1 online).
Figure 2.
Subcellular Localization of YFP-PMD1 and YFP-PMD2.
Confocal images are from leaf epidermal cells from transgenic plants expressing PMD1pro:YFP-PMD1 (A) or 35Spro:YFP-PMD2 (B) along with the peroxisomal marker CFP-PTS1 or the mitochondrial marker COX4-CFP. YFP signals are in green, and CFP signals are in magenta. Merged images show the colocalization of the YFP fusion protein to peroxisomes or mitochondria. Bars = 5 μm.
Although PMD2 was not found to be associated with peroxisomes in our initial transient studies, the sequence and structural similarities it shares with PMD1 prompted us to reanalyze its subcellular localization. To this end, YFP-PMD2 was expressed under the control of the 35S promoter in Arabidopsis plants that carry CFP-PTS1 or COX4-CFP. Confocal imaging of T2 transgenic plants discovered the localization of YFP-PMD2 on the surface of COX4-CFP–labeled mitochondria, as shown by ring-like structures outside mitochondria. However, none of the YFP-PMD2 signals colocalized with the CFP-PTS1–marked peroxisomes, suggesting that the protein is sorted to mitochondria only (Figure 2B).
In summary, our data corroborated with results from a recent genome-wide analysis of C-TA proteins in Arabidopsis, which showed the localization of PMD1 to mitochondria (Kriechbaumer et al., 2009). In addition, we uncovered the association of PMD1 with peroxisomes. By contrast, PMD1’s homolog, PMD2, localizes exclusively to mitochondria.
PMD1 Is C-Terminally Anchored to the Membrane of Peroxisomes and Mitochondria
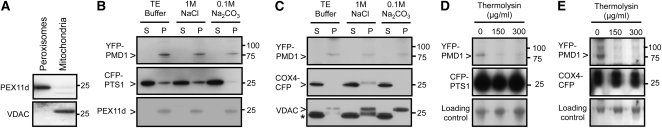
A biochemical approach was employed to verify the presence of PMD1 on peroxisomes and mitochondria. We first isolated peroxisomes and mitochondria from leaves of 4-week-old Arabidopsis transgenic plants expressing 35Spro:YFP-PMD1 using previously published methods (Kruft et al., 2001; Werhahn and Braun, 2002; Reumann et al., 2009). To determine the purity of the isolated organelles, ~10 μg of each type of organelle proteins was separated on SDS-PAGE gels (see Supplemental Figure 2 online) and then subjected to immunoblot analyses using organelle-specific antibodies (i.e., α-PEX11d [Peroxin 11d] for peroxisomes and α-VDAC [voltage-dependent anion channel protein] for mitochondria). Our data showed that the peroxisomal and mitochondrial proteins were highly enriched (Figure 3A). The peroxisomal and mitochondrial proteins were then treated by Tris-EDTA buffer, a high concentration of sodium chloride (1 M NaCl), and strong alkaline solutions (Na2CO3, pH 11.0), respectively, to separate membrane from soluble proteins and integral membrane proteins from those that are peripherally associated with the membrane. Soluble and insoluble fractions were separated through centrifugation. Similar to the peroxisomal membrane protein PEX11d and the mitochondrial membrane protein VDAC, YFP-PMD1 was detected only in the insoluble fractions of both peroxisomal and mitochondrial proteins after the treatments (Figures 3B and 3C), suggesting that PMD1 is embedded in the membrane of the organelles. By contrast, the peroxisomal matrix protein CFP-PTS1 and the mitochondrial luminal protein COX4-CFP were mostly detected in the soluble fractions (Figures 3B and 3C). We concluded that PMD1 is an integral membrane protein of the peroxisome and mitochondrion.
Figure 3.
PMD1 Is Anchored to the Organelle Membranes by the C Terminus.
(A) Immunoblotting analysis of peroxisomal and mitochondrial proteins purified from Arabidopsis plants expressing YFP-PMD1 using antibodies against the peroxisomal specific protein PEX11d and the mitochondrial-specific protein VDAC.
(B) and (C) Immunoblot analyses of purified Arabidopsis leaf peroxisomes (B) and mitochondria (C) after the proteins were treated with sodium chloride (NaCl) or sodium carbonate (Na2CO3) and fractionated into soluble (S) and pellet (P) fractions. The GFP antibody was used to detect the expression of CFP and YFP fusion proteins. CFP-PTS1 and COX4-CFP served as controls for matrix proteins, and PEX11d and VDAC were organelle-specific membrane protein controls. Asterisk in (C) marks cross-hybridized signals of COX4-CFP, as the same membrane had been probed by the GFP antibody in the previous immunoblot.
(D) and (E) Immunoblot analyses of peroxisomes (D) and mitochondria (E) that were treated with various concentrations of thermolysin. The levels of CFP and YFP fusion proteins were detected with the GFP antibody. A nonspecific band was used as loading control.
Numbers to the right of each panel indicate molecular mass in kilodaltons.
Previously characterized C-TA proteins share a similar membrane topology on the subcellular compartments to which they are tethered; that is, a cytosolic N terminus that contains the functional domain(s), a TMD at or close to the C terminus, and a short C-terminal end protruding into the matrix/luminal side of the compartments (Borgese et al., 2007; Abell and Mullen, 2011). To determine the topology of PMD1 on the organelle membranes, we performed protease protection assays, in which we treated the purified peroxisomes and mitochondria with thermolysin, a protease that can only access and degrade the cytosolic part of the outer membrane proteins (Tranel et al., 1995). Since YFP was fused to the N-terminal end of PMD1, a green fluorescent protein (GFP) antibody was used to probe the long CC domain that was hypothesized to be exposed to the cytosolic side of the organelle membranes. Immunoblot analysis with α-GFP demonstrated that, after thermolysin treatments, YFP-PMD1 was barely detected, whereas the amount of the matrix/lumenal proteins CFP-PTS1 and COX4-CFP remained largely unchanged (Figures 3D and 3E). Our data collectively demonstrated that PMD1 is anchored to the membrane of peroxisomes and the outer envelope membrane of mitochondria by its C terminus, with the N-terminal long CC domain facing the cytosol.
Mutants of PMD1 Exhibit Abnormal Morphologies and Abundance of Peroxisomes and Mitochondria
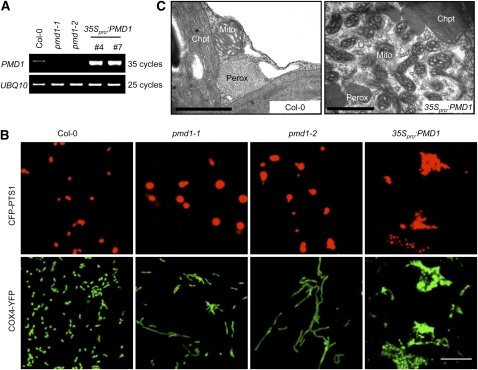
The identification of the dual-localized plant-specific protein PMD1 prompted us to determine whether this protein plays a role in regulating the morphology, size, and/or abundance of peroxisomes and mitochondria. To this end, we characterized two T-DNA insertion alleles of PMD1: pmd1-1 (CS84214), which has a T-DNA insertion in the middle of the coding region, and pmd1-2 (SALK_139577), which has an insertion 60 bp upstream from the translational start site ATG (Figure 1A). RT-PCR analyses of RNA from the T-DNA insertion lines did not detect PMD1 transcripts in either allele (Figure 4A), indicating that, despite their overall indistinguishable appearance from the wild type throughout development (see Supplemental Figure 3A online), pmd1-1 and pmd1-2 are both null mutants. We then transformed the peroxisomal (CFP-PTS1) and the mitochondrial (COX4-YFP) markers into the mutants to examine the organelle morphologies. Confocal images taken from T2 plants revealed that both pmd1-1 and pmd1-2 contain peroxisomes that are larger in diameter than those in the wild type (Figure 4B). A quantification of the size and abundance of peroxisomes showed that the average size of a peroxisome in the pmd1 mutants, as measured by CFP fluorescent area using Image J, is ~2.7 times that of a peroxisome in the wild type, whereas the total numbers of peroxisomes reduced to 65 to 70% in the mutant (see Supplemental Figures 4A and 4B online). Mitochondria in pmd1-1 and pmd1-2 were markedly elongated (Figure 4B). The average size of a mitochondrion increased ~2.5 to 2.7 times, and the total number of mitochondria decreased to ~64 to 72% of that in the wild-type plants (see Supplemental Figures 4C and 4D online). The organelle phenotypes of the loss-of-function pmd1 mutants pointed to a role of the PMD1 protein in the morphogenesis and possibly division of peroxisomes and mitochondria.
Figure 4.
pmd1 Mutants Exhibit Abnormal Peroxisomal and Mitochondrial Morphologies.
(A) RT-PCR analyses showing the levels of PMD1 mRNA in the wild type (Col-0), loss-of-function mutants (pmd1-1 and pmd1-2), and gain-of-function mutants (35Spro:PMD1 line numbers 4 and 7). UBQ10 transcripts are used as loading controls.
(B) Confocal images from leaf epidermal cells of the indicated plants showing morphologies of peroxisomes (labeled by CFP-PTS1) and mitochondria (marked by COX4-YFP). Bar = 10 μm.
(C) TEM images from leaf mesophyll cells illustrating the ultrastructure of peroxisomes (Perox), mitochondria (Mito), and chloroplasts (Chpt) in Col-0 and 35Spro:PMD1 plants. Bars = 1 μm.
[See online article for color version of this figure.]
To further study the function of PMD1 in the organelles, we ectopically expressed PMD1 in wild-type Columbia-0 (Col-0) Arabidopsis plants containing both CFP-PTS1 and COX4-YFP and confirmed the induction of the genes in the transgenic plants by RT-PCR (Figure 4A). Plants expressing 35Spro:PMD1 displayed retarded growth (see Supplemental Figure 3A online) and massive proliferation and aggregation of peroxisomes and mitochondria as observed by confocal microscopy (Figure 4B). Transmission electron microscopy (TEM) analysis further revealed that the aggregated organelles were composed of overproliferated peroxisomes and mitochondria, which are smaller than those in the wild type (Figure 4C). Within a 2-μm2 area of a leaf mesophyll cell, we normally found one to two peroxisomes or mitochondria in a wild-type plant, yet as many as five peroxisomes and up to 12 mitochondria were present in the 35Spro:PMD1 plants. Likewise, plants expressing 35Spro:YFP-PMD1 also displayed increased proliferation and aggregation of peroxisomes and mitochondria and inhibition of plant growth (see Supplemental Figures 3B and 3C online). The organelle phenotypes in the PMD1-overexpressing plants, together with phenotypes observed in the pmd1 null mutants, suggested a positive role for PMD1 in increasing the abundance of peroxisomes and mitochondria.
PMD1 Interacts with Itself but Lacks Physical Interaction with Known Peroxisomal and Mitochondrial Division Factors in Arabidopsis
Organelle phenotypes of the PMD1 gain- and loss-of-function mutants are to a large extent reminiscent of those of FIS1, which encodes a dual-localized C-TA protein serving as the membrane anchor for DRPs (reviewed in Kaur and Hu, 2009). In light of this, we speculated that PMD1 may be a plant-specific factor with a role similar to FIS1 in the fission of peroxisomes and mitochondria, possibly by functioning in the same complex as FIS1 and DRP3. In addition, since CC proteins are well known for their ability to interact with themselves or other CC domain–containing proteins (Rose et al., 2004, 2005), we also wanted to determine whether PMD1 can form homocomplexes through self-interaction. To this end, we conducted coimmunoprecipitation (co-IP) and yeast two-hybrid (Y2H) assays to test whether PMD1 can interact or form a complex with itself, FIS1, and DRP.
For the co-IP assay, YFP was fused to the N-terminal end of the full-length coding sequences of PMD1, DRP3A, DRP3B, DRP5B, FIS1A, and FIS1B, respectively, to create bait proteins. We also generated the prey protein biotinylated-PMD1 (BIO-PMD1) by fusing the biotin carboxyl carrier protein domain of the biotinylate subunit of MCCase (MCCA; At1g03090) (Qi and Katagiri, 2009) to the N terminus of PMD1. Each YFP- and BIO-fusion protein pair was then transiently coexpressed in tobacco leaves. Proteins extracted from the infiltrated leaves were subjected to immuno-pull down by agarose-conjugated GFP antibody, followed by immunoblot analysis. As shown in Supplemental Figure 5A online, most of the YFP fusion proteins were expressed at very low levels in the input samples, unable to be detected by α-GFP. However, they were highly enriched by immunoprecipitation, suggesting that they are efficiently pulled down by α-GFP. Consistent with the notion that CC proteins dimerize, BIO-PMD1 was pulled down by YFP-PMD1, whereas the control protein YFP-PTS1 was unable to precipitate BIO-PMD1, suggesting that PMD1 interacts with itself. Similar to YFP-PTS1, none of the YFP-DRP and YFP-FIS1 proteins were able to pull down BIO-PMD1 (see Supplemental Figure 5A online), which is indicative of a lack of physical interaction or close proximity between PMD1 and the Arabidopsis DRP3A, DRP3B, DRP5B, and FIS1 proteins.
We used Y2H assays (Proquest two-hybrid system) to verify the co-IP results. PMD1 deleted for TMD was cloned into pDestTM32 to generate PMD1ΔTMD-DNA binding domain fusion. PMD1ΔTMD, DRP3/DRP5B, and FIS1ΔTMD were cloned into pDESTTM22 to create protein fusions with the activation domain. The proteins were then coexpressed in yeast strain Mav203. A robust growth of cells on medium lacking Ura, Trp, and His in the presence of 10 mM 3-amino-1,2,4-triazole suggested an interaction between the two proteins expressed. Consistent with our co-IP data, only a strong self-interaction of PMD1 was detected (see Supplemental Figure 5B online). Taken together, results from the Y2H assays supported our conclusion that PMD1 can interact with itself, yet it has no physical interaction or does not form a complex with the known fission factors DRP3A, DRP3B, DRP5B, or FIS1.
Although physical interactions between PMD1 and DRP3s were not detected by co-IP and Y2H experiments, the membrane-anchored PMD1 protein may still be indirectly involved in the recruitment of DRP3s to the division sites of peroxisomes and mitochondria. To test this possibility, we used the Fast Agrobacterium-mediated Seedling Transformation technique (Li et al., 2009) to transiently express 35Spro:YFP-DRP3A or 35Spro:YFP-DRP3B in wild-type Col-0 or pmd1-1 seedlings, which are already expressing the peroxisomal marker CFP-PTS1 or the mitochondrial marker COX4-CFP. Both YFP-DRP3A and YFP-DRP3B were properly targeted to the constriction sites or tips of peroxisomes and mitochondria in pmd1-1, just like in the wild-type cells (see Supplemental Figure 6 online), suggesting that PMD1 alone is not critical for recruiting DRP3 proteins to the organelles. Results from the co-IP, Y2H, and protein localization studies collectively led us to the conclusion that PMD1 may mediate peroxisomal and mitochondrial division/proliferation by a mechanism that is independent from the action of the FIS1-DRP3 complex.
PMD1 Interacts with PMD2, Which Is Specifically Involved in Mitochondrial Morphogenesis
The lack of association of PMD1 with known division factors led us to take alternative approaches to identify PMD1-interacting proteins, with the hope to uncover the functional role of PMD1 in organelle proliferation. For this purpose, we adopted two independent affinity purification methods, biotin-streptavidin purification (Qi and Katagiri, 2009) and GFP purification. To perform biotin-streptavidin purification, PMD1pro:BIO-PMD1 was expressed in mcca (SALK_137966) background to eliminate the purification of endogenous MCCA (Qi and Katagiri, 2009). To pull down proteins interacting with YFP-PMD1, we used plants expressing 35Spro:YFP-PMD1 and CFP-PTS1. Arabidopsis plants expressing the fusion proteins were subjected to biotin-streptavidin purification as described by Qi and Katagiri (2009) or GFP pull-down assays (see Methods), and mcca and plants expressing YFP-PTS1 alone were used as negative controls. The purified proteins were separated on SDS-PAGE and visualized by silver staining (see Supplemental Figures 7A and 7B online). Proteins pulled down by each strategy were identified using liquid chromatography–tandem mass spectrometry as previously described (Reumann et al., 2009), with two replicates for each method. PMD2 was the only protein identified in common by both methods, suggesting that PMD2 might be a bona fide PMD1-interacting protein.
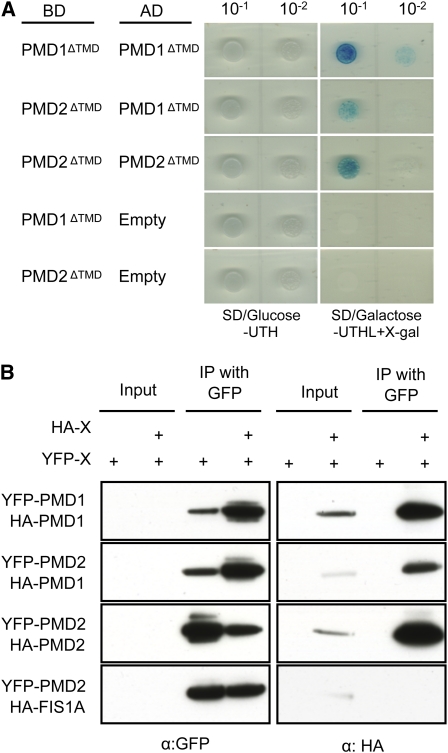
We confirmed the interaction between PMD1 and PMD2 using Y2H and co-IP assays. Interaction between PMD1 and PMD2, both of which were deleted for the putative TMD, was first tested using the Matchmaker LexA Y2H system. When the protein pairs PMD1ΔTMD and PMD1ΔTMD, PMD1ΔTMD and PMD2ΔTMD, and PMD2ΔTMD and PMD2ΔTMD were coexpressed in the yeast strain EGY48, yeast cells grew strongly in medium lacking Ura, Trp, His, and Leu (SD/galactose-UTHL) and turned blue in the presence of 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside. By contrast, yeast cells transformed with the PMD1ΔTMD or PMD2ΔTMD fusion protein and an empty vector did not grow on the selection media, indicating specific interactions between the tested PMD protein pairs (Figure 5A). We next performed co-IP to confirm the Y2H results, using YFP- and hemagglutinin (HA)–tagged PMD proteins transiently expressed in tobacco leaf epidermal cells. YFP-PMD1 and YFP-PMD2 efficiently pulled down HA-PMD1 and HA-PMD2, respectively, and YFP-PMD2 pulled down HA-PMD1 (Figure 5B). Consistent with the lack of interaction between PMD1 and FIS1 (see Supplemental Figure 5 online), YFP-PMD2 also failed to pull down HA-FIS1A (Figure 5B). These data demonstrated the ability for PMD1 and PMD2 to form homo- as well as heterocomplexes.
Figure 5.
PMD1 and PMD2 Are Able to Form Homo- and Heterocomplexes.
(A) Y2H analyses. SD/Glc-UTH selects for transformants and SD/Gal-UTHL + X-Gal selects for protein–protein interactions. Empty, pB42AD-GW vector only.
(B) Co-IP analyses. Various combinations of the fusions as indicated were transiently expressed in tobacco leaf epidermal cells, followed by immunoprecipitation using the GFP antibody. A GFP or HA antibody was used to detect the proteins.
[See online article for color version of this figure.]
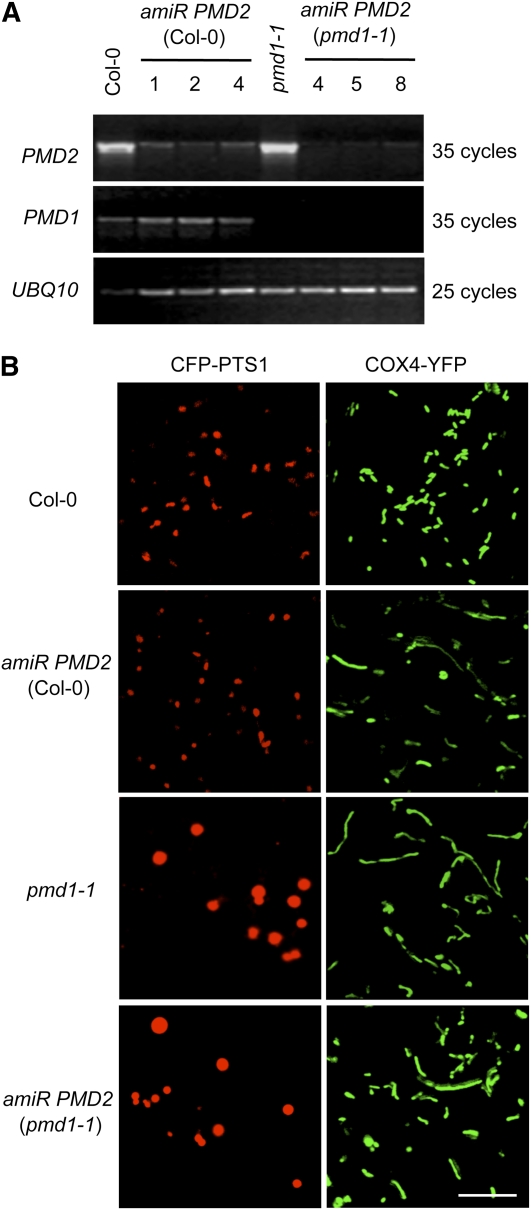
To characterize the functional role of PMD2, it was necessary to observe organelle morphologies in loss-of-function pmd2 mutants. Since T-DNA insertion mutants for PMD2 were unavailable, we generated artificial microRNA (amiR) lines (see Methods and Supplemental Figure 7C online) to specifically reduce the expression of PMD2 in wild-type Col-0 plants, which expressed the peroxisomal marker CFP-PTS1 and the mitochondrial marker COX4-YFP. In addition, we transformed amiR PMD2 into pmd1-1 plants, which also coexpressed the peroxisomal and mitochondrial fluorescent markers, to see whether compounded phenotypes can be created in the double mutant. We obtained 16 transgenic amiR PMD2 lines each in the Col-0 and pmd1-1 background. RT-PCR analysis showed efficient knockdown of the expression of PMD2 in 15 lines in the Col-0 background, without affecting the transcript level of PMD1, and in 14 lines in the pmd1-1 background. Results from three lines in each background, all of which were indistinguishable from wild-type plants in appearance, are presented in Figure 6A.
Figure 6.
PMD2 Is Involved in Mitochondrial Morphogenesis.
(A) RT-PCR analyses showing the transcripts of PMD1, PMD2, and UBQ10 in Col-0, amiR PMD2, pmd1-1, and pmd1-1 amiR PMD2 plants. Three independent lines that contain amiR PMD2 in the Col-0 (lines 1, 2, and 4) or pmd1-1 (lines 4, 5, and 8) background are shown.
(B) Confocal images from leaf epidermal cells of plants coexpressing the organelle markers. Peroxisomes were labeled by CFP-PTS1, and mitochondria were marked by COX4-YFP. Bar = 10 μm.
[See online article for color version of this figure.]
Plants from the T3 generation of the PMD2 knockdown lines were analyzed by confocal microscopy to observe the peroxisomal and mitochondrial morphologies. Consistent with PMD2’s specific localization to mitochondria (Figure 2B), suppression of the PMD2 gene rendered morphological changes to mitochondria only (Figure 6B). The size and number of mitochondria in the amiR PMD2 lines were comparable to those in the pmd1 mutants (see Supplemental Figures 4C and 4D online). On the other hand, no obvious effect on the morphology and abundance of peroxisomes was observed (Figure 6B; see Supplemental Figures 4A and 4B online). The double mutant contained peroxisomes with morphology and size similar to those in pmd1; its mitochondrial phenotype was also similar to those in pmd1 or amiR PMD2 (Figure 6B; see Supplemental Figure 4 online). We concluded that PMD2 has an exclusive role in the morphogenesis and/or proliferation of mitochondria, and the functions of PMD1 and PMD2 in mitochondria are nonredundant.
The C Termini of the PMD Proteins Contain Organelle Targeting Signals
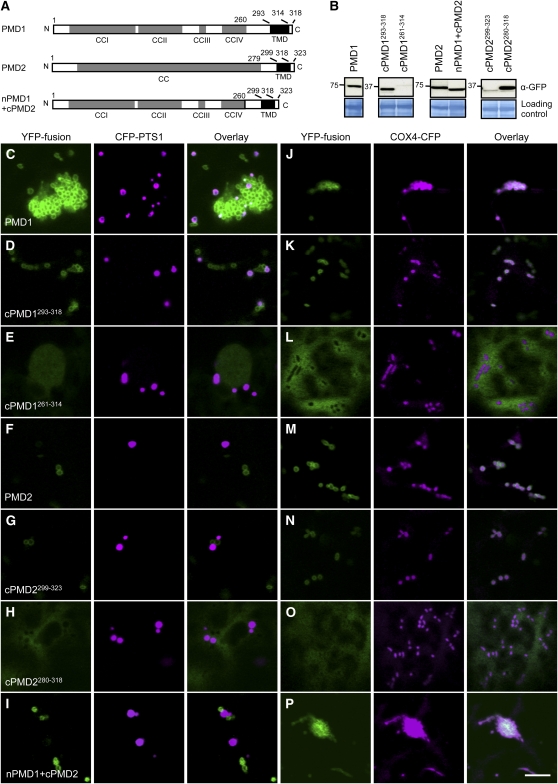
Previous studies of C-TA proteins demonstrated that their organelle-specific targeting signals reside in the TMD and its flanking sequences (reviewed in Abell and Mullen, 2011). Despite their sequence similarities along the length of the proteins, PMD1 and PMD2 showed distinct organelle targeting patterns (i.e., PMD1 is dual targeted, whereas PMD2 only targets to mitochondria). To determine whether the C terminus carries targeting signals for PMD1 and PMD2, we made various truncations of the PMD proteins, which were subsequently fused to the C terminus of YFP. The 35S promoter–driven fusion proteins were transiently expressed in tobacco leaves together with the peroxisomal marker CFP-PTS1 or the mitochondrial marker COX4-CFP (Figures 7A and 7B; see Supplemental Figures 8A and 8B online).
Figure 7.
Organelle Targeting Signals Reside in the C Terminus of PMD1 and PMD2.
(A) Schematics of PMD1, PMD2, and nPMD1+cPMD2 with the CC domains, TMD, and amino acids indicated. Despite sequence similarities between PMD1 and PMD2, PMD2 was annotated as having a single and long CC domain.
(B) Immunoblot analysis to detect the YFP-PMD variants expressed in tobacco cells. The large subunit of ribulose-1,5-bis-phosphate carboxylase/oxygenase was used as the loading control. Numbers to the left of each panel indicate molecular mass in kilodaltons.
(C) to (P) Subcellular targeting of the YFP-PMD1/PMD2 fusions. YFP signals are in green, and CFP signals are in magenta. To better illustrate colocalization between the YFP fusion and CFP-PTS1, images in (C) were taken from a region where peroxisome proliferation/aggregation was not so strong. Bar = 10 μm.
Confocal microscopy of tobacco leaf epidermal cells showed that, like full-length PMD1, the C terminus of PMD1 (cPMD1261-318 and cPMD1293-318) was able to direct the YFP fusion protein to both peroxisomes and mitochondria, whereas YFP fused to the N terminus of PMD1 (nPMD11-260) mislocalized to the cytosol (Figures 7C, 7D, 7J, and 7K; see Supplemental Figures 8C, 8D, 8G, and 8H online). To determine whether the sequence downstream from TMD is required for organelle targeting, we also tested the localization of YFP-cPMD1261-314, in which the last four amino acids were deleted, and found the fusion proteins to be distributed in the cytosol (Figures 7E and 7L). These data together suggested that the segment covering TMD and the C-terminal end 3′ to TMD is necessary and sufficient for PMD1’s dual targeting. Furthermore, although ectopically expressed YFP-PMD1 often led to organelle proliferation in tobacco cells (Figures 7C and 7J), as it did in transgenic Arabidopsis plants (see Supplemental Figure 3B online), overexpression of YFP-cPMD1 did not cause such a phenotype (Figures 7D and 7K; see Supplemental Figures 8D and 8H online). This result supports the view that the functional domain of PMD1 is located at its cytoplasmic N terminus.
Similar to what was found for PMD1, the C terminus, but not the N terminus, of PMD2 was critical to direct the YFP to mitochondria (Figures 7F, 7G, 7M, and 7N; see Supplemental Figures 8E, 8F, 8I, and 8J online). In addition, the five amino acids downstream from the TMD at the C-terminal end are also required for PMD2’s localization to mitochondria, as YFP-cPMD2280-318 localized to the cytosol (Figures 7H and 7O). Thus, like many previously reported C-TA proteins, PMD1 and PMD2 both use the C terminus, which includes the TMD and the sequences 3′ to the TMD, for their organelle targeting. However, unlike YFP-PMD1, overexpression of YFP-PMD2 did not cause mitochondrial overproliferation in tobacco cells (Figures 7F and 7M) or transgenic Arabidopsis plants (Figure 2B). These findings suggest that PMD2’s role in mitochondrial morphogenesis may be distinct from that of PMD1. This notion is further supported by the fact that a chimeric protein, in which the N terminus of PMD1 was fused to the C terminus of PMD2 (nPMD1+cPMD2; Figure 7A), exclusively localized to mitochondria and induced massive mitochondrial proliferation (Figure 7P). We concluded that the PMD proteins carry their organelle targeting signals at the C termini and the functional domains at the N termini. For PMD1 in particular, its N terminus is responsible for inducing the proliferation of the organelles.
DISCUSSION
PMD1 Is a Plant-Specific Protein That Promotes Peroxisomal/Mitochondrial Proliferation Independently from the FIS1-DRP3 Complex
The peroxisomal and mitochondrial division machineries are composed of evolutionarily conserved factors, such as DRP and FIS1, as well as lineage-specific proteins, such as Mdv1p and Caf4p from yeast, Mff1 from metazoans, and the mitochondrial division protein ELM1 from Arabidopsis (see Introduction). In this work, we discovered PMD1 as another plant-specific factor involved in the organelle division/proliferation process. Null mutants of PMD1 have enlarged peroxisomes and elongated mitochondria, whereas overexpression of the gene leads to massive proliferation and aggregation of these organelles (Figure 4). These phenotypes are to large degrees similar to what we previously observed in the mutants of Arabidopsis FIS1, which encode a C-TA protein that is dual localized to peroxisomes and mitochondria and presumably recruits DRP proteins to the division site. The loss-of-function fis1A and fis1B mutants contain enlarged peroxisomes and mitochondria, and the FIS1 overexpressors have an increased number of both organelles, which are often aggregated together (Scott et al., 2006; Zhang and Hu, 2008; Zhang and Hu, 2009). Results from the mutant analyses in this work suggest that PMD1 is involved in the division/proliferation of peroxisomes and mitochondria in Arabidopsis.
Among the lineage-specific peroxisomal and mitochondrial division proteins, Mdv1p and Caf4p are part of the FIS1-DRP complex. Mff1 and ELM1 both function independently from FIS1, yet are still required for the recruitment of DRP proteins (reviewed in Kaur and Hu, 2009). By contrast, PMD1 lacks physical interaction with FIS1 or DRP3/DRP5B in both co-IP and Y2H analyses (see Supplemental Figure 5 online). It does not play an obvious role in the organelle targeting of DRP3A and DRP3B either (see Supplemental Figure 6 online). Based on these data, we speculate that PMD1 directly or indirectly mediates the division/proliferation of peroxisomes and mitochondria independently from the FIS1 and DRP3/DRP5B proteins.
Loss-of-function mutants of PMD1, PMD2, and the pmd1 pmd2 double mutant do not show obvious defects in growth and development. PMD1 overexpressors are slow growing (see Supplemental Figure 3 online), and this growth defect might be a consequence of the gross organelle aggregation. It seems that although PMD1 and PMD2 have obvious functions in organelle morphogenesis, loss of these proteins is not sufficient to severely disturb the physiology of the organelles under general plant growth conditions. The fis1A fis1B double mutant is also slightly impaired in growth (Zhang and Hu, 2009). It would be interesting to generate a mutant in which the functions of all four C-TA proteins, PMD1, PMD2, FIS1A, and FIS1B, are knocked out to determine whether the pathways, in which PMD1/PMD2 and FIS1A/FIS1B exert their functions, are partially redundant. The mutants can also be challenged with various stresses to determine whether one of the pathways operates primarily under some adverse growth conditions.
PMD1 and PMD2 Play Nonredundant Roles
Although PMD1 and PMD2 share 35% amino acid identity and similar domain structures, these two proteins are not redundant in function. First, PMD1 is dual localized and mediates the morphogenesis and proliferation of both peroxisomes and mitochondria. However, PMD2 appears to have an exclusive function in the morphogenesis of mitochondria. In addition, the fact that YFP-PMD2 forms a ring-like pattern outside mitochondria and that PMD2 and PMD1 share similar domain structures strongly suggest that PMD2 is another C-TA protein that is tethered to the outer envelope membrane of mitochondria. However, the functions of PMD1 and PMD2 in mitochondria do not overlap. The amiR PMD2 lines, in which the expression of PMD2 is significantly reduced, phenocopy the elongated mitochondrial phenotype of pmd1-1, suggesting that PMD2 possesses a similar molecular function as PMD1 on mitochondria. However, the pmd1-1 amiR PMD2 double mutant displays a mitochondrial phenotype identical to the pmd single mutants, leading us to speculate that the functions of these two proteins in mitochondria are not exchangeable. Third, unlike PMD1, overexpression of PMD2 does not induce mitochondrial proliferation, further supporting the view that these two proteins carry out distinct functions in mitochondria. Based on these findings, we favor the hypothesis that PMD1 and PMD2 form a complex on the membrane of mitochondria and function cooperatively to mediate the morphogenesis and/or proliferation of mitochondria, with PMD1 playing a rate-limiting role.
Mode of Action for the PMD Proteins
Eukaryotic genomes contain a large number of genes encoding C-TA proteins that are involved in various cellular processes, from gene expression to vesicle trafficking (Abell and Mullen, 2011). Bioinformatic screens predicted the presence of >500 C-TA proteins in Arabidopsis; the functions of the majority of them are unknown (Kriechbaumer et al., 2009; Pedrazzini, 2009). The functional domains in the C-TA proteins often reside in the N terminus, which occupies the bulk of the protein. For PMD1, its N-terminal region has a long CC domain that can be separated into four shorter CC domains (Figure 1).
CC proteins are ubiquitous eukaryotic proteins with diverse functions. Whereas short CC domains often function as dimerization motifs in transcription factors, other CC domains, including those that are >100 amino acids long, have been found to be involved in the attachment of proteins or protein complexes to larger subcellular compartments (Gillingham and Munro, 2003; Rose et al., 2005). Given that several membrane-bound CC proteins in organelle division serve as membrane tethers for effector proteins (see Introduction) and the phenotypes of the pmd1 mutants, the most likely function for PMD1 is to recruit downstream cytosolic effectors for division. The lack of interaction detected between PMD1 and the peroxisomal/mitochondrial division effector DRP3 implies that other yet unidentified effector proteins may be recruited by PMD1 to the organelle membranes.
Our confocal microscopy analysis showed that overexpressing the PMD1 gene causes massive clustering/aggregation of peroxisomes and mitochondria (Figure 4), hinting at a role for PMD1 in organelle positioning and distribution. However, further TEM experiments revealed more and smaller peroxisomes and mitochondria in the overexpressors, suggesting a role of PMD1 in organelle division/proliferation rather than distribution. The clustering phenotype may have been caused by the fact that the division effect of ectopically expressed PMD1 overwhelmed the machinery responsible for separating the organelles after division. Consistent with this hypothesis, cells overexpressing Arabidopsis FIS1A or FIS1B also show peroxisomal and mitochondrial aggregations (Zhang and Hu, 2008). Given that PMD1 forms homocomplexes, the strong organelle aggregations may also have been caused by dimerization of the overly abundant membrane-tethered PMD1 proteins between organelles.
Although we favor a role for PMD1 (and likely PMD2 as well) in inducing the proliferation/division of the organelles, other possibilities also exist. For example, they may be involved in shaping of peroxisomes and mitochondria with a yet unknown mechanism, which indirectly affects the abundance of these organelles. In addition, although little is known about the fusion of peroxisomes and mitochondria in plants, it is also a formal possibility that PMD proteins function directly as suppressors of organelle fusion. Lastly, given the organelle aggregation phenotype in the PMD1 overexpressors and the fact that PMD1 and PMD2 form homo- and heterocomplexes, it was tempting to hypothesize that these proteins may function as molecular tethers in maintaining a close proximity/juxtaposition of the metabolically connected peroxisomes and mitochondria. Supporting this hypothesis is the findings that some CC-containing proteins are involved in tethering mitochondria to the ER in mice (de Brito and Scorrano, 2008) or binding the vacuole to mitochondria in Cyanidoschyzon merolae (Fujiwara et al., 2010). However, so far we have been unable to observe differences in the distance/association between peroxisomes and mitochondria in the pmd single and double mutants and wild-type plants, thus making this hypothesis less favored at present.
Organelle Targeting Mechanism for PMD1 and PMD2
In this work, we provided evidence to demonstrate PMD1’s dual targeting to peroxisomes and mitochondria and PMD2’s exclusive localization to mitochondria. At this point, we do not have evidence for preferential targeting of PMD1 to a particular organelle, as almost all CFP-PTS1–labeled peroxisomes and COX4-CFP–labeled mitochondria colocalized with YFP-PMD1. However, in general we do observe fewer peroxisomes than mitochondria in a given cell. In our experience, it is easier to capture high-quality images of mitochondria near the surface of cells, where much fewer peroxisomes are found.
Our biochemical analysis demonstrated that YFP-PMD1 is an integral membrane protein on both organelles. In the membrane association assay, the YFP-PMD1 protein bands detected in the mitochondrial fraction is much weaker than those in the peroxisomal proteins (Figures 3B and 3C). One possible explanation is that YFP-PMD1 is not as tightly associated with the mitochondrial membrane as it is to the peroxisomal membrane and therefore was easier to be lost during organelle isolation procedures. Alternatively, given that the proteome of mitochondria is 10 times larger than that of the peroxisome (Reumann et al., 2004; Millar et al., 2008), there is a higher enrichment of peroxisomal proteins than mitochondrial proteins when an equal amount of organelle proteins are used in the assays and, hence, the peroxisome-localized protein shows a stronger band on the immunoblot.
Multiple pathways have been reported on the sorting of C-TA proteins to their destined cellular membranes; various pathways even exist for proteins targeting to the same membrane, such as the ER membrane (Abell and Mullen, 2011). Given the small number of plant C-TA proteins that have been characterized, especially those localized to peroxisomes, and the complex nature of the targeting mechanisms, predicting the targeting destinations of C-TA proteins with high accuracy may not be a simple task (Kriechbaumer et al., 2009). Consistent with the view that the targeting signals for C-TA proteins are located at the C termini, we have shown in this study that the C-terminal tail of PMD1 or PMD2, which possesses the putative TMD and the flanking sequences downstream of TMD, is necessary and sufficient to target the proteins to their destined organelles. The peroxisomal targeting of the human C-TA proteins FIS1 (hFis1) and PEX26 requires the cytoplasmic receptor/chaperone protein PEX19; binding sites for PEX19 were also mapped onto the C terminus of these proteins (Halbach et al., 2006; Delille and Schrader, 2008). The C-terminal sequences of these proteins and PMD1/PMD2 vary greatly; therefore, it is hard to predict the role for the Arabidopsis PEX19 homolog in PMD1’s peroxisomal targeting without experimental evidence. Taken together, a much more detailed dissection of this region in PMD1 and PMD2 will be needed to pinpoint precisely the residues and structural features required for peroxisomal or mitochondrial targeting and to further identify factors that mediate the targeting.
Peroxisomes and Mitochondria Are Interconnected in Various Ways
In plants, an ever-increasing number of proteins have been found to be dual localized to peroxisomes and mitochondria, raising the interesting possibility that these two organelles are more closely connected than previously known. Our discovery of PMD1 as a dual-localized protein that mediates the division/proliferation of both peroxisomes and mitochondria further substantiates this notion.
In addition to the metabolic pathways and division factors that link peroxisomes and mitochondria, other ways of interaction or communication also exist between these two organelles. For instance, in yeast, mitochondrial dysfunction induces peroxisome biogenesis and increases peroxisomal functions via a retrograde signaling pathway controlled by the transcription factor RTG (Chelstowska and Butow, 1995; Epstein et al., 2001). In addition, a mammalian RIG-I-like receptor adaptor protein called MADS, which is also a C-TA protein, was recently found to be anchored to the membrane of peroxisomes and the outer envelope membrane of mitochondria, serving as part of an antiviral signaling system to induce the expression of defense genes (Seth et al., 2005; Dixit et al., 2010). Furthermore, mammalian mitochondrial-derived vesicles carrying specific cargo were found to merge with a population of peroxisomes. Interestingly, the vesicles carry the MAPL protein, a SUMO ligase that can enhance the activity of the mitochondrial/peroxisomal division protein Drp1 (Neuspiel et al., 2008), providing evidence for a new way of communication between mitochondria and peroxisomes (Andrade-Navarro et al., 2009). Our study supports the view that subcellular compartments within a eukaryotic cell are highly interactive. In particular, the regulation of the abundance, morphology, and distribution of the metabolically related peroxisomes and mitochondria may be highly coordinated to maintain cellular homeostasis. Further investigation of the role of the PMD proteins and other dual-targeted proteins will be instrumental to a better understanding of the coordination and communication among cellular compartments.
METHODS
Plant Material, Growth Conditions, Transformation, and Plant Selection
Arabidopsis thaliana plants were grown at 20°C with 70% humidity and irradiated with 70 to 80 μmol m−2 s−2 of white light for 14 h per day. T-DNA insertion mutants, pmd1-1 (CS84214) and pmd1-2 (SALK_139577), were obtained from the ABRC (Columbus, OH). pmd1-1 is in Col-3 background and pmd1-2 is in Col-0 background. The presence of the T-DNAs and the homozygosity of mutants were identified by PCR of genomic DNA using the following primers: pmd1-1, CS854214-LP, CS854214-RP, Wisc LP; pmd1-2, SALK_139577-LP, SALK_139577-RP, Lbbnew. The absence of PMD1 transcripts was determined by RT-PCR using PMD1-attB1 and PMD1-attB2-N as listed in Supplemental Table 2 online.
To generate plants with organelle markers, the peroxisomal marker CFP-PTS1 (conferring resistance to gentamycin) and the mitochondrial marker Saccharomyces cerevisiae COX4-YFP were cotransformed into the null mutants or wild-type Col-0. The COX4-YFP construct has two versions: one confers resistance to kanamycin and was used for transformation into pmd1-1 and Col-0, and the other confers resistance to Basta and was used for transformation of pmd1-2. T1 generation was screened with kanamycin or basta for the presence of the mitochondrial marker and with an epifluorescence microscope for the presence of CFP signals (peroxisomes) and YFP signals (mitochondria). The T2 or T3 generation of the transgenic plants was subjected to confocal microscopy to identify plants that contain both markers.
To generate gain-of-function mutants, 35Spro:PMD1 or 35Spro:YFP-PMD1/PMD2 was constructed as described below and transformed into the double marker plants in Col-0 background. Transgenic plants were screened with Basta, and the T3 generations were subjected to confocal imaging. To generate knockdown mutants, amiR PMD2 was constructed as described below and transformed into the double marker plant in Col-0 or pmd1-1 background. The transgenic plants were screened with hygromycin, and the T3 generation was used to observe organelle and plant morphologies.
The simplified Arabidopsis transformation method (http://www.plantpath.wisc.edu/fac/afb/protocol.html) was used to perform all Arabidopsis plant transformations using Agrobacterium tumefaciens strain GV3101 (pMP90). For selections of transgenic plants, T1 seeds were plated on 0.5 Linsmaier and Skoog medium with 100 μg/mL of gentamycin, 50 μg/mL of kanamycin, or 50 μg/mL of hygromycin. For Basta screening, T1 individuals were grown on soil and sprayed at 7 and 9 d after germination with 0.1% (v/v) of Basta (Finale; Farnam Companies) and 0.025% (v/v) Silwet L-77 to select transgenic plants. The putative transgenic plants were further screened with epifluorescence microscopy, which confirmed that the transgenes fused with a fluorescent protein tag were expressed, or with RT-PCR, which checked the expression levels of the overexpressed genes without a fluorescent protein tag.
A. tumefaciens–mediated transient expression in Arabidopsis was performed according to a published method (Li et al., 2009). Four-day-old Arabidopsis Col-0 and pmd1-1 expressing the peroxisomal marker CFP-PTS1 or the mitochondrial marker COX4-CFP were inoculated with A. tumefaciens cells harboring YFP-DRP3A or YFP-DRP3B to transiently express the fusion genes. Two days later, the transfected plants were subjected to confocal imaging.
Tobacco (Nicotiana tabacum) plants were grown at 24°C with 70% humidity and irradiated with 50 μmol m−2 s−2 of white light for 14 h per day. Six- to eight-week-old plants were subjected to A. tumefaciens–mediated infiltration to transiently express the genes of interest. A. tumefaciens cells harboring the plasmid(s) of interest were incubated at 28°C with shaking (200 rpm) for ~24 h. The cells were then spun down, washed, and resuspended with water to an A600 of 0.05. Cells harboring genes of interest were infiltrated into mature leaves using a syringe. The infiltrated plants were incubated in the same growth condition for 2 d. Then, the infiltrated leaves were subjected to confocal imaging, co-IP, or immunoblot analysis.
Gene Cloning and Plasmid Construction
The genes of interest were amplified with Gateway-compatible primers from the cDNA synthesized from total RNA of wild-type (Col-0) seedlings or cDNA clones obtained from the ABRC using Phusion High-Fidelity DNA polymerase (New England Biolabs). The PCR fragments were cloned to the donor vector (pDonor 207 or pDonor/Zeo) and different destination vectors using a standard Gateway cloning system (Invitrogen). PMD1pro:YFP-PMD1 was generated using an overlapping PCR approach (http://gfp.stanford.edu/protocol/index5.html) and cloned into a binary vector pGWB1 (Nakagawa et al., 2007). To generate YFP fusion proteins, the genes of interest were cloned into the binary vector pEarleyGate 104 (Earley et al., 2006) or pDest-35S-6xHis-YFP (kindly provided by Sheng Quan, Michigan State University). To clone overexpressors, the genes were cloned into pEarleyGate 100 (Earley et al., 2006) or pGWB2 (Nakagawa et al., 2007).
To create the biotinylated PMD1 construct, a biotin tag containing 80 amino acids from the biotin carboxyl carrier protein domain of the Arabidopsis MCCA gene (At1g03090) was fused to the N terminus of PMD1 using the overlapping PCR method described above. The overlapped PCR fragment was subcloned into pMDC32 (CD3-738) (Qi and Katagiri, 2009). Later, the 35S promoter was replaced with the PMD1 native promoter using restriction sites KpnI and HindIII.
To create HA fusion proteins, the full-length coding sequence of PMD1, PMD2, and FIS1 was cloned into the binary vector pEarleyGate 201 (Earley et al., 2006).
To clone the amiRNA construct, the WMD3-Web microRNA designer (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi) was used to design amiRNA PMD2. One of the recommended amiRNAs, 5′-(489) TGACAGTCCTTAAACCAGCGC (509)-3′, was selected and cloned using an overlapping PCR as described in the WMD3-Web microRNA designer (http://wmd3.weigelworld.org/downloads/Cloning_of_artificial_microRNAs.pdf). The precursor miRNA was amplified by Gateway-compatible primers and cloned into pGWB2 (Nakagawa et al., 2007). The primers and vectors used in this study are listed in Supplemental Tables 2 and 3 online.
RT-PCR Analysis
RNA from leaves of 4-week-old Arabidopsis plants were purified as previously described (Zhang and Hu, 2009). For RT-PCR analysis, 0.5 μg of total RNA was used to make cDNA using high-capacity RNA-to-cDNA master mix (Applied Biosystems). PCR amplification was performed as previously described (Zhang and Hu, 2010) using primers specific for PMD1 (At3g58840), PMD2 (At1g06530), and UBQ10 (At4g05320). The number of cycles used in the RT-PCR was optimized to be in the linear range of amplification. Primers used in this study are listed in Supplemental Table 2 online.
Immunoblot Analysis
Fifty milligrams of fresh tissue was ground with plastic pestles using liquid nitrogen and 500 μL of SDS-containing extraction buffer (60 mM Tris-HCl, pH 8.8, 2% SDS, 2.5% glycerol, 0.13 mM EDTA, pH 8.0, and 1× protease inhibitor cocktail complete from Roche). The tissue lysates were vortexed for 30 s, heated at 70°C for 10 min, and centrifuged at 13,000g twice for 5 min at room temperature. The supernatants were then transferred to new tubes. For SDS-PAGE analysis, 5 μL of the extract in 1× NuPAGE LDS sample buffer (Invitrogen) was separated on 4 to 12% NuPage (Invitrogen) before being transferred to the polyvinylidene fluoride membrane. The membrane was incubated with 3% BSA in 1× TBST (50 mM Tris-base, 150 mM NaCl, and 0.05% Tween 20, pH 8.0) overnight at 4°C. Then, it was probed with the antibody prepared in the blocking buffer at room temperature for 1 h. The antibodies used are as follows: 1:20,000 α-GFP (Abcam), 1:20,000 α-horseradish peroxidase–conjugated streptavidin (Millipore), 1:1000 α-PEX11d (Orth et al., 2007), 1:5000 α-VDAC (Reumann et al., 2009), and 1:100 α-HA (Cell Signaling). The probed membrane was washed three times with 1× TBST for 5 min before incubation with the secondary antibody at room temperature for 1 h. The secondary antibodies used are 1:20,000 goat anti-rabbit IgG for α-GFP and α-PEX11d, 1:20,000 goat anti-mouse IgG for α-horseradish peroxidase–conjugated Streptavidin (Millipore), α-VDAC, and α-HA. Finally, the membrane was washed four times with 1× TBST for 10 min before the signals were visualized with SuperSignal West Dura Extended duration substrate (Pierce Biotechnology).
Purification of Peroxisomal and Mitochondrial Proteins
Rosette leaves from 4-week-old transgenic plants expressing 35Spro:YFP-PMD1 and an organelle marker were used for organelle purification. Peroxisomes were isolated from the peroxisomal marker (CFP-PTS1) background as described previously (Reumann et al., 2009).
Mitochondria were purified from the mitochondrial marker (COX4-CFP) background as described (Kruft et al., 2001; Werhahn and Braun, 2002) with minor modifications. Leaves were harvested and homogenized on ice with 120 mL of grinding buffer (450 mM Suc, 1.5 mM EGTA, 0.2% BSA, 0.6% polyvinylpyrrolidone-40, 10 mM DTT, 0.2 mM phenylmethylsulfonyl fluoride, and 15 mM MOPS/KOH, pH 7.4) using a mortar and a pestle. The homogenized solution was filtered with two layers of Miracloth. Chloroplasts and other organelles and particles were sedimented by centrifugation for 10 min at 3500g and 5 min at 6000g. The supernatant was then centrifuged for 10 min at 17,000g to pellet down the fraction enriched in mitochondria. The pellet was washed with 80 mL of washing buffer (0.3 M Suc and 10 mM MOPS/KOH, pH 7.2). Ten milliliters of mitochondria in washing buffer was topped onto a three-step Percoll gradient, which contains 18, 29, and 45% Percoll in 0.3 M Suc and 10 mM MOPS/KOH, pH 7.2. The Percoll gradients were centrifuged for 40 min at 45,000g; mitochondria were recovered from the interphase between 29 and 45%. The mitochondrial fractions were pooled together and diluted with washing buffer. After two washes with washing buffer and a 10-min centrifugation at 17,000g for 10 min, the mitochondria were resuspended with 3 mL of washing buffer containing the protease inhibitor cocktail complete (Roche).
The purity of peroxisomal and mitochondrial proteins was determined using immunoblot analyses as described above. A polyclonal PEX11d antiserum, which was raised against Arabidopsis PEX11d (Orth et al., 2007), was used to detect the expression of the peroxisome-specific protein PEX11d. A monoclonal VDAC antiserum, which was raised against the VDAC protein of maize (Zea mays) and used in our previous study (Reumann et al., 2009), was used to detect the mitochondrion-specific protein VDAC.
Assays for Membrane Association and Topology
Peroxisomal and mitochondrial membrane association of PMD1 was tested as previously described (Orth et al., 2007). The orientation of the CC domains was tested by protease protection using thermolysin as described previously (Cline et al., 1984) with minor modifications. Purified peroxisomal and mitochondrial proteins (200 μL) were treated with 0, 150, or 300 μg/mL of thermolysin in an incubation buffer containing 50 mM HEPES/NaOH, pH 7.5, 0.33 M sorbitol, and 0.5 mM CaCl2. The reactions were performed at 4°C for 1 h and stopped by adding 5 mM EDTA. Twenty microliters of the treated proteins were subjected to immunoblot analysis using the GFP antibody as described above.
Co-IP Assay
BIO-PMD1, YFP-PMD1/PMD2, and HA-PMD1/PMD2/FIS1A proteins were transiently expressed in tobacco as described above. Approximately 1 g fresh weight of infiltrated leave was collected 2 d after infiltration. The tissue was homogenized in RIPA buffer (Thermo) with 1× complete protease inhibitor cocktail (Roche) and lysed on a rotator at 4°C for 1 h. The samples were centrifuged at 13,000g for 10 min to remove cell debris. The supernatants were then incubated with 20 μL of agarose-conjugated anti-GFP (MBL) on a rotator for 1 h to pull down the YFP fusion proteins. The agarose beads were then spun down at 3000g for 15 s and washed four times with RIPA buffer. Proteins associated with the YFP fusion protein were eluted by adding 50 μL of 1× NuPAGE LDS sample buffer (Invitrogen) and heating at 75°C for 10 min. The eluted proteins were analyzed by immunoblot assays as described above.
Y2H Assays
The ProQuest two-hybrid system was used to test the interaction between PMD1 and peroxisome division factors. PMD1ΔTMD was cloned into pDest32 to generate the bait clone. Full-length DRP, FIS1ΔTMD, and PMD1ΔTMD were cloned into the pDest22 vector to generate the prey clones. The plasmid DNA was transformed into yeast strain MaV203. The presence of the transformed plasmid DNAs was screened using standard synthetic dropout medium (SD/-Ura-Trp). SD medium with 10 mM 3-amino-1,2,4-triazole and without Ura, Trp, and His was used to test the physical interaction of the tested proteins.
The Matchmaker two-hybrid system (Clontech) was also used to test the self-interaction of PMD1 and PMD2 as well as the interaction between PMD1 and PMD2. PMDΔTMD was cloned into a derivative of pGilda or pB42AD containing the Gateway cassette (attR1-Cmr-ccdB-arrR2). The plasmid DNA was transformed into the yeast strain EGY48 using the Frozen-EZ yeast transformation kit as recommended by the manufacturer (Zymo). Transformants were screened using standard synthetic dropout medium (SD/Glc-Ura-Trp-His). SD/Gal medium with X-Gal and minus Ura, Trp, His, and Leu was used to test for protein interaction.
Confocal and TEM Analyses
All the confocal images were taken by a confocal laser scanning microscope (Zeiss LSM 510 META). Before imaging, leaf discs were fixed with a fixation buffer, which contains 2% formaldehyde, 10 mM EGTA, 5 mM MgSO4, and 50 mM PIPES, pH 7.0, for 10 min to stop the movement of organelles and then rinsed with water. YFP and CFP signals were detected as previously reported (Reumann et al., 2009). All images, except those shown in Figures 4B and 6B, were obtained from a single focal plane. For images in Figures 4B and 6B, a Z series of six images, each 10 μm in depth, was collected and superimposed into one image.
To observe the ultrastructure of organelles in mesophyll cells, 4-week-old wild-type (Col-0) and 35Spro:PMD1 Arabidopsis plants were subjected to TEM analysis using methods previously described (Duek and Fankhauser, 2005).
Quantification of Organelles
Peroxisomes and mitochondria were visualized in leaf epidermal cells from the wild type and various pmd mutants that express the fluorescent organelle markers using confocal microscopy. A Z series of 10 images, each 18 μm in depth, was collected and superimposed into one image to quantify peroxisomes. A total of 10 images from the wild type and mutants was analyzed by Image J (http://rsb.info.nih.gov/ij) as previously reported (Desai and Hu, 2008). The size of peroxisomes or mitochondria was measured as average fluorescent area per organelle using units assigned by Image J. The P values were calculated by Student's two-tailed t test against the wild type.
Affinity Purification of the PMD1 Protein Complex
To perform the biotin-streptavidin purification, T3 homozygous plants expressing cMCCA-PMD1 were used to isolate the PMD1-interacting proteins using methods described before (Qi and Katagiri, 2009). For the GFP pull-down assay, T3 homozygous plants expressing YFP-PMD1 in the peroxisomal marker (CFP-PTS1) and mitochondrial marker (COX4-CFP) backgrounds were used. Thirty grams of leaf tissues was homogenized in 50 mL of RIPA buffer (Thermo) containing 1× complete protease inhibitor cocktail (Roche) and lysed on a rotator at 4°C for 1 h. The samples were subjected to centrifugation at 13,000g for 10 min to remove cell debris. The supernatants were then incubated with 200 μL of agarose-conjugated anti-GFP (MBL) on a rotator for 1 h to pull down the YFP fusion proteins. The agarose beads were spun down at 3000g for 15 s and washed four times with RIPA buffer. The YFP-interacting proteins were eluted by adding 1× NuPAGE LDS sample buffer (Invitrogen) and heating at 75°C for 10 min. The eluted proteins were subjected to electrophoresis in 4 to 12% gradient NuPAGE gels (Invitrogen) and silver staining or liquid chromatography–tandem mass spectrometry analyses as described before (Reumann et al., 2009).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: PMD1 (At3g58840), PMD2 (At1g06530), UBQ10 (At4g05320), DRP3A (At4g33650), DRP3B (At2g14120), DRP5B (At3g19720), FIS1A (At3g57090), and FIS1B (At5g12390). Germplasm identification numbers for pmd1-1 and pmd1-2 alleles used in this works are as follows: pmd1-1 (WiscDsLox393-396O15; The Arabidopsis Information Resource stock number CS854214) and pmd1-2 (SALK_139577).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. C-Terminal YFP Fusion of PMD1 (PMD1-YFP, in Green) Is Localized to the Cytosol.
Supplemental Figure 2. Proteins from Purified Peroxisomes and Mitochondria.
Supplemental Figure 3. Plant Morphologies of the pmd Loss- and Gain-of-Function Lines and Localization of YFP-PMD1 in Transgenic Plants.
Supplemental Figure 4. Quantification of Organelle Size and Number.
Supplemental Figure 5. PMD1 Self-Interacts but Does Not Interact with the Known Peroxisomal and Mitochondrial Division Factors.
Supplemental Figure 6. Localization of DRP3 Proteins in the pmd1 Mutant.
Supplemental Figure 7. Proteins Pulled Down from 35Spro:YFP-PMD1 or PMD1pro:BIO-PMD1 Plants and the Gene Structure of PMD2.
Supplemental Figure 8. Additional Data to Figure 7 on the Identification of the Peroxisomal and Mitochondrial Targeting Signals on PMD1 and PMD2.
Supplemental Table 1. In Silico Identification of Peroxisomal Membrane Proteins Containing Long Coiled-Coil Domains.
Supplemental Table 2. Primers Used in This Study.
Supplemental Table 3. Vectors Used in This Study.
Acknowledgments
We thank the ABRC (Columbus, OH) for providing the T-DNA insertion mutants, Detlef Weigel (Max Planck Institute for Developmental Biology, Tübingen, Germany) for providing the pRS300 vector for amiRNA cloning, and Tsuyoshi Nakagawa (Research Institute of Molecular Genetics, Shimane University, Matsue, Japan) for the pGWB1 and pGWB2 vectors. We also thank Melinda Frame and Alicia Pastor (Center of Advanced Microscopy, Michigan State University) for their help with confocal microscopy and TEM analyses. This work was supported by grants from the National Science Foundation (MCB 0618335) and the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (DE-FG02-91ER20021) to J.H.
AUTHOR CONTRIBUTIONS
K.A. designed and performed the experiments, analyzed the data, and wrote the first draft of the article. J.H. contributed to experimental design, data analysis, and writing of the article.
References
- Abell B.M., Mullen R.T. (2011). Tail-anchored membrane proteins: exploring the complex diversity of tail-anchored-protein targeting in plant cells. Plant Cell Rep. 30: 137–151 [DOI] [PubMed] [Google Scholar]
- Andrade-Navarro M.A., Sanchez-Pulido L., McBride H.M. (2009). Mitochondrial vesicles: An ancient process providing new links to peroxisomes. Curr. Opin. Cell Biol. 21: 560–567 [DOI] [PubMed] [Google Scholar]
- Arimura S., Aida G.P., Fujimoto M., Nakazono M., Tsutsumi N. (2004). Arabidopsis dynamin-like protein 2a (ADL2a), like ADL2b, is involved in plant mitochondrial division. Plant Cell Physiol. 45: 236–242 [DOI] [PubMed] [Google Scholar]
- Arimura S., Fujimoto M., Doniwa Y., Kadoya N., Nakazono M., Sakamoto W., Tsutsumi N. (2008). Arabidopsis ELONGATED MITOCHONDRIA1 is required for localization of DYNAMIN-RELATED PROTEIN3A to mitochondrial fission sites. Plant Cell 20: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura S., Tsutsumi N. (2002). A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division. Proc. Natl. Acad. Sci. USA 99: 5727–5731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung K., Hu J. (2009). The Arabidopsis peroxisome division mutant pdd2 is defective in the DYNAMIN-RELATED PROTEIN3A (DRP3A) gene. Plant Signal. Behav. 4: 542–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker A., Graham I.A., Holdsworth M., Smith S.M., Theodoulou F.L. (2006). Chewing the fat: Beta-oxidation in signalling and development. Trends Plant Sci. 11: 124–132 [DOI] [PubMed] [Google Scholar]
- Borgese N., Brambillasca S., Colombo S. (2007). How tails guide tail-anchored proteins to their destinations. Curr. Opin. Cell Biol. 19: 368–375 [DOI] [PubMed] [Google Scholar]
- Chelstowska A., Butow R.A. (1995). RTG genes in yeast that function in communication between mitochondria and the nucleus are also required for expression of genes encoding peroxisomal proteins. J. Biol. Chem. 270: 18141–18146 [DOI] [PubMed] [Google Scholar]
- Cline K., Werner-Washburne M., Andrews J., Keegstra K. (1984). Thermolysin is a suitable protease for probing the surface of intact pea chloroplasts. Plant Physiol. 75: 675–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brito O.M., Scorrano L. (2008). Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456: 605–610 [DOI] [PubMed] [Google Scholar]
- Delille H.K., Alves R., Schrader M. (2009). Biogenesis of peroxisomes and mitochondria: Linked by division. Histochem. Cell Biol. 131: 441–446 [DOI] [PubMed] [Google Scholar]
- Delille H.K., Schrader M. (2008). Targeting of hFis1 to peroxisomes is mediated by Pex19p. J. Biol. Chem. 283: 31107–31115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M., Hu J. (2008). Light induces peroxisome proliferation in Arabidopsis seedlings through the photoreceptor phytochrome A, the transcription factor HY5 HOMOLOG, and the peroxisomal protein PEROXIN11b. Plant Physiol. 146: 1117–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit E., et al. (2010). Peroxisomes are signaling platforms for antiviral innate immunity. Cell 141: 668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek P.D., Fankhauser C. (2005). bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci. 10: 51–54 [DOI] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Epstein C.B., Waddle J.A., Hale W., IV, Davé V., Thornton J., Macatee T.L., Garner H.R., Butow R.A. (2001). Genome-wide responses to mitochondrial dysfunction. Mol. Biol. Cell 12: 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasanu A., Fagarasanu M., Rachubinski R.A. (2007). Maintaining peroxisome populations: A story of division and inheritance. Annu. Rev. Cell Dev. Biol. 23: 321–344 [DOI] [PubMed] [Google Scholar]
- Fan J., Quan S., Orth T., Awai C., Chory J., Hu J. (2005). The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol. 139: 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M., Arimura S., Mano S., Kondo M., Saito C., Ueda T., Nakazono M., Nakano A., Nishimura M., Tsutsumi N. (2009). Arabidopsis dynamin-related proteins DRP3A and DRP3B are functionally redundant in mitochondrial fission, but have distinct roles in peroxisomal fission. Plant J. 58: 388–400 [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Kuroiwa H., Yagisawa F., Ohnuma M., Yoshida Y., Yoshida M., Nishida K., Misumi O., Watanabe S., Tanaka K., Kuroiwa T. (2010). The coiled-coil protein VIG1 is essential for tethering vacuoles to mitochondria during vacuole inheritance of Cyanidioschyzon merolae. Plant Cell 22: 772–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldón T., Snel B., van Zimmeren F., Hemrika W., Tabak H., Huynen M.A. (2006). Origin and evolution of the peroxisomal proteome. Biol. Direct 1: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandre-Babbe S., van der Bliek A.M. (2008). The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell 19: 2402–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Kadirjan-Kalbach D., Froehlich J.E., Osteryoung K.W. (2003). ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc. Natl. Acad. Sci. USA 100: 4328–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham A.K., Munro S. (2003). Long coiled-coil proteins and membrane traffic. Biochim. Biophys. Acta 1641: 71–85 [DOI] [PubMed] [Google Scholar]
- Halbach A., Landgraf C., Lorenzen S., Rosenkranz K., Volkmer-Engert R., Erdmann R., Rottensteiner H. (2006). Targeting of the tail-anchored peroxisomal membrane proteins PEX26 and PEX15 occurs through C-terminal PEX19-binding sites. J. Cell Sci. 119: 2508–2517 [DOI] [PubMed] [Google Scholar]
- Hoepfner D., Schildknegt D., Braakman I., Philippsen P., Tabak H.F. (2005). Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122: 85–95 [DOI] [PubMed] [Google Scholar]
- Kaur N., Hu J. (2009). Dynamics of peroxisome abundance: A tale of division and proliferation. Curr. Opin. Plant Biol. 12: 781–788 [DOI] [PubMed] [Google Scholar]
- Kaur N., Reumann S., Hu J. (2009). Peroxisome biogenesis and function. In The Arabidopsis Book 7: e0113, doi/10.1199/tab.0123
- Kobayashi S., Tanaka A., Fujiki Y. (2007). Fis1, DLP1, and Pex11p coordinately regulate peroxisome morphogenesis. Exp. Cell Res. 313: 1675–1686 [DOI] [PubMed] [Google Scholar]
- Koch A., Thiemann M., Grabenbauer M., Yoon Y., McNiven M.A., Schrader M. (2003). Dynamin-like protein 1 is involved in peroxisomal fission. J. Biol. Chem. 278: 8597–8605 [DOI] [PubMed] [Google Scholar]
- Koch A., Yoon Y., Bonekamp N.A., McNiven M.A., Schrader M. (2005). A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell 16: 5077–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriechbaumer V., Shaw R., Mukherjee J., Bowsher C.G., Harrison A.M., Abell B.M. (2009). Subcellular distribution of tail-anchored proteins in Arabidopsis. Traffic 10: 1753–1764 [DOI] [PubMed] [Google Scholar]
- Kruft V., Eubel H., Jänsch L., Werhahn W., Braun H.P. (2001). Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol. 127: 1694–1710 [PMC free article] [PubMed] [Google Scholar]
- Li J.F., Park E., von Arnim A.G., Nebenführ A. (2009). The FAST technique: A simplified Agrobacterium-based transformation method for transient gene expression analysis in seedlings of Arabidopsis and other plant species. Plant Methods 5: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingard M.J., Gidda S.K., Bingham S., Rothstein S.J., Mullen R.T., Trelease R.N. (2008). Arabidopsis PEROXIN11c-e, FISSION1b, and DYNAMIN-RELATED PROTEIN3A cooperate in cell cycle-associated replication of peroxisomes. Plant Cell 20: 1567–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Rost B. (2001). Comparing function and structure between entire proteomes. Protein Sci. 10: 1970–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano S., Nakamori C., Kondo M., Hayashi M., Nishimura M. (2004). An Arabidopsis dynamin-related protein, DRP3A, controls both peroxisomal and mitochondrial division. Plant J. 38: 487–498 [DOI] [PubMed] [Google Scholar]
- Millar A.H., Small I.D., Day D.A., Whelan J. (2008). Mitochondrial biogenesis and function in Arabidopsis. In The Arabidopsis Book 6: e0111, doi/10.1199/tab.0105 [DOI] [PMC free article] [PubMed]
- Miyagishima S.Y., Froehlich J.E., Osteryoung K.W. (2006). PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell 18: 2517–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A.M., Ward G.P., Hettema E.H. (2008). Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J. Cell Sci. 121: 1633–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy A.D., McCaffery J.M., Shaw J.M. (2000). Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151: 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagotu S., Krikken A.M., Otzen M., Kiel J.A., Veenhuis M., van der Klei I.J. (2008). Peroxisome fission in Hansenula polymorpha requires Mdv1 and Fis1, two proteins also involved in mitochondrial fission. Traffic 9: 1471–1484 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Neuspiel M., Schauss A.C., Braschi E., Zunino R., Rippstein P., Rachubinski R.A., Andrade-Navarro M.A., McBride H.M. (2008). Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr. Biol. 18: 102–108 [DOI] [PubMed] [Google Scholar]
- Orth T., Reumann S., Zhang X., Fan J., Wenzel D., Quan S., Hu J. (2007). The PEROXIN11 protein family controls peroxisome proliferation in Arabidopsis. Plant Cell 19: 333–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung K.W., Nunnari J. (2003). The division of endosymbiotic organelles. Science 302: 1698–1704 [DOI] [PubMed] [Google Scholar]
- Otera H., Wang C., Cleland M.M., Setoguchi K., Yokota S., Youle R.J., Mihara K. (2010). Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 191: 1141–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrazzini E. (2009). Tail-anchored proteins in plants. J. Plant Biol. 52: 1226–1239 [Google Scholar]
- Penfield S., Pinfield-Wells H.M., Graham I. (2006). Storage reserve mobilisation and seedling establishment in Arabidopsis. In The Arabidopsis Book 4: e0100, doi/10.1199/tab.0100 [DOI] [PMC free article] [PubMed]
- Peterhansel C., Horst I., Niessen M., Blume C., Kebeish R., Kurkcuoglu S., Kreuzaler F. (2010). Photorespiration. In The Arabidopsis Book 8: e0130, doi/10.1199/tab.0130 [DOI] [PMC free article] [PubMed]
- Praefcke G.J., McMahon H.T. (2004). The dynamin superfamily: Universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 5: 133–147 [DOI] [PubMed] [Google Scholar]
- Purdue P.E., Lazarow P.B. (2001). Peroxisome biogenesis. Annu. Rev. Cell Dev. Biol. 17: 701–752 [DOI] [PubMed] [Google Scholar]
- Qi Y., Katagiri F. (2009). Purification of low-abundance Arabidopsis plasma-membrane protein complexes and identification of candidate components. Plant J. 57: 932–944 [DOI] [PubMed] [Google Scholar]
- Reumann S., Ma C., Lemke S., Babujee L. (2004). AraPerox. A database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol. 136: 2587–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Quan S., Aung K., Yang P., Manandhar-Shrestha K., Holbrook D., Linka N., Switzenberg R., Wilkerson C.G., Weber A.P., Olsen L.J., Hu J. (2009). In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol. 150: 125–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A., Manikantan S., Schraegle S.J., Maloy M.A., Stahlberg E.A., Meier I. (2004). Genome-wide identification of Arabidopsis coiled-coil proteins and establishment of the ARABI-COIL database. Plant Physiol. 134: 927–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A., Schraegle S.J., Stahlberg E.A., Meier I. (2005). Coiled-coil protein composition of 22 proteomes—Differences and common themes in subcellular infrastructure and traffic control. BMC Evol. Biol. 5: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter A., Fourcade S., Ripp R., Mandel J.L., Poch O., Pujol A. (2006). The evolutionary origin of peroxisomes: An ER-peroxisome connection. Mol. Biol. Evol. 23: 838–845 [DOI] [PubMed] [Google Scholar]
- Scott I., Tobin A.K., Logan D.C. (2006). BIGYIN, an orthologue of human and yeast FIS1 genes functions in the control of mitochondrial size and number in Arabidopsis thaliana. J. Exp. Bot. 57: 1275–1280 [DOI] [PubMed] [Google Scholar]
- Seth R.B., Sun L., Ea C.K., Chen Z.J. (2005). Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122: 669–682 [DOI] [PubMed] [Google Scholar]
- Tieu Q., Nunnari J. (2000). Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 151: 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu Q., Okreglak V., Naylor K., Nunnari J. (2002). The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J. Cell Biol. 158: 445–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko V.I., Mullen R.T. (2006). Peroxisome biogenesis: the peroxisomal endomembrane system and the role of the ER. J. Cell Biol. 174: 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranel P.J., Froehlich J., Goyal A., Keegstra K. (1995). A component of the chloroplastic protein import apparatus is targeted to the outer envelope membrane via a novel pathway. EMBO J. 14: 2436–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders R.J., Waterham H.R. (2006). Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 75: 295–332 [DOI] [PubMed] [Google Scholar]
- Werhahn W., Braun H.P. (2002). Biochemical dissection of the mitochondrial proteome from Arabidopsis thaliana by three-dimensional gel electrophoresis. Electrophoresis 23: 640–646 [DOI] [PubMed] [Google Scholar]
- Yang Y., Glynn J.M., Olson B.J., Schmitz A.J., Osteryoung K.W. (2008). Plastid division: Across time and space. Curr. Opin. Plant Biol. 11: 577–584 [DOI] [PubMed] [Google Scholar]
- Zhang X., Hu J. (2009). Two small protein families, DYNAMIN-RELATED PROTEIN3 and FISSION1, are required for peroxisome fission in Arabidopsis. Plant J. 57: 146–159 [DOI] [PubMed] [Google Scholar]
- Zhang X., Hu J. (2010). The Arabidopsis chloroplast division protein DYNAMIN-RELATED PROTEIN5B also mediates peroxisome division. Plant Cell 22: 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.C., Hu J.P. (2008). FISSION1A and FISSION1B proteins mediate the fission of peroxisomes and mitochondria in Arabidopsis. Mol. Plant 1: 1036–1047 [DOI] [PubMed] [Google Scholar]