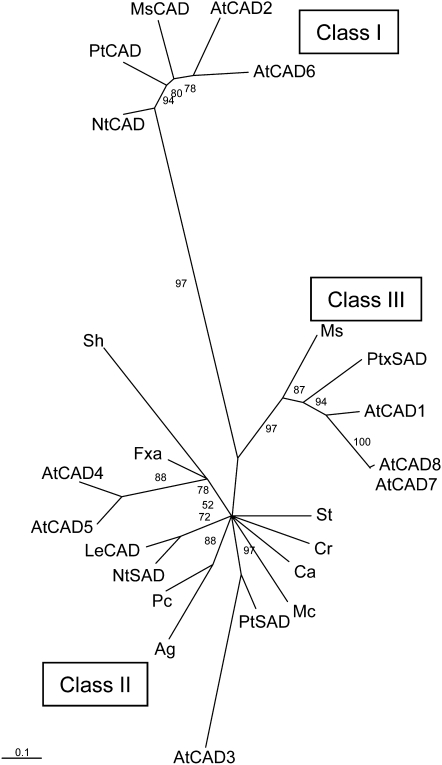
Figure 2.
Phylogenetic Tree of CAD and SAD Proteins.
CAD and SAD protein sequences were aligned using Clustal W (see Supplemental Data Set 1 online), and a phylogenetic tree was produced using a maximum likelihood method (Tree-Puzzle 5.0) with quartet puzzling support values. The amino acid substitution matrix used was JTT. Rate heterogeneity among sites was modeled using a gamma distribution with four categories. The tree was viewed in Tree-View v.1.6.6. The three major clades, denoted Class I, Class II, and Class III CADs by Raes et al. (2003),are indicated. Arabidopsis CAD genes are named according to Raes et al. (2003). Ag, Apium graveolens; At, Arabidopsis thaliana; Ca, Camptotheca acuminate; Cr, Catharanthus roseus; Fxa, Fragaria × ananassa; Le, Solanum lycopersicum; Mc, Mesembryanthemum crystallinum; Ms Medicago sativa; Nt, Nicotiana tabacum; Pc, Petroselinum crispum; Pt, Populus tremuloides; Ptx, Populus tremula × Populus tremuloides; Sh, Stylosanthes humilis; St, Solanum tuberosum.
Bar = 0.1 amino acid substitutions.