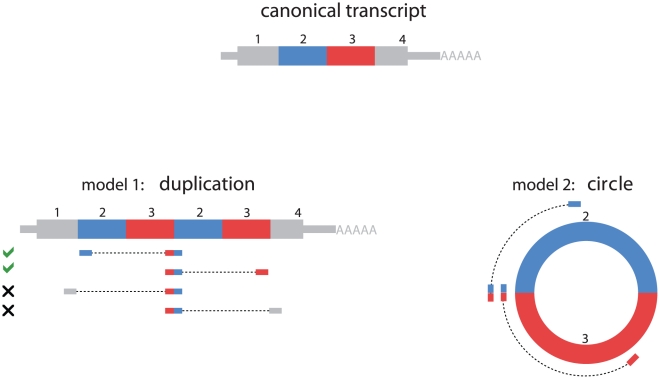
Figure 1. Models to explain exon scrambling.
The canonical linear reference transcript is depicted with exons as colored boxes with four exons 1, 2, 3, and 4. Two simple models of RNA structure that could explain scrambled transcripts are depicted at left and right. At left, model 1 depicts how a scrambled exon 3-exon 2 junction could arise from a tandem duplication of exons 3 and 2, positioning the first copy of exon 3 upstream of exon 2. At the RNA level, this event could arise from post-transcriptional exon rearrangement, or a genomic duplication of exons 2 and 3. Under the model of tandem duplication, when one side of a paired-end read maps to the junction between exon 3 and 2, the other may map to any of exons 1, 2, 3 or 4 with probabilities determined by the library's insert length distribution and the exon lengths. Our data supports paired-end mapping between a junction and exons 2 or 3, but not exons 1 and 4. We note that in principle, the scrambled exon 3 - exon 2 junction could arise from other splicing events and does not necessarily entail tandem duplication. At right, model 2 depicts how a scrambled exon 3 - exon 2 junction could arise from splicing of exons 2 and 3 into a circular RNA molecule, again positioning exon 3 upstream of exon 2. In this model, when one side of a paired-end read maps to the junction between exon 3 and 2, the other will map to exon 2 or exon 3.