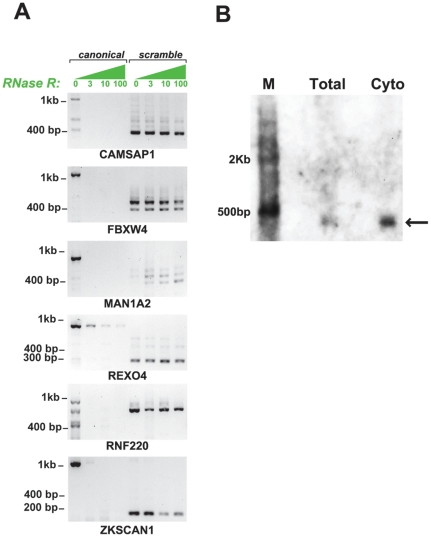
Figure 3. RNaseR assay confirms scrambled exons arise from circular RNA.
Panel A: Total RNA from HeLa cells was digested with RNaseR at varying enzyme concentrations (0, 3, 10, and 100 units) after the RNA was depleted of ribosomal RNA. Primers capable of amplifying the canonical linear transcript and the predicted circular transcript (by outward facing primers within a single exon predicted in the scramble) were used in a RT-PCR experiment for each of the digestion conditions. Canonical transcripts were consistently degraded by RNaseR, only detectable by PCR at 0 units of RNaseR, whereas predicted circular transcripts consistently resisted the RNaseR challenge, providing strong evidence of circularity. FBXW4 and MAN1A2 respectively show 2 and 4 circular isoforms, both of which were predicted by the sequencing data. The predicted lengths of circular isoforms are respectively a 3-2 junction of CAMSAP1 (predicted to produce a 435 bp circle), a 4-2 and 5-2 junction of FBXW4 (predicted to produce 415 and 510 bp circles), a 4-2, 5-2 and 6-2 junction of MAN1A2 (predicted to produce 471, 553, and 648 bp circles), a 3-3 junction in REXO4 (predicted to produce a 338 bp circle), a 2-2 junction of RNF220 (predicted to produce a 742 bp circle) and a 3-2 junction of ZKSCAN1 (predicted to produce a 667 bp circle). Panel B: A northern blot on total and cytoplasmic lysate from HeLa cells shows hybridization of a 481 bp probe complementary to the MAN1A2 5-2 exon scramble. 3.7 and 6.2 ug of total and cytoplasmic RNA were loaded onto a 1% agarose gel and 10 pM of probe was hybridized for 24–48 hours. Detection was performed using the BrightStar BioDetect Kit (Ambion, Austin, TX). The specific band at 553 bp corresponds to the predicted size of a circular RNA containing exons 2,3,4 and 5 of MAN1A2.