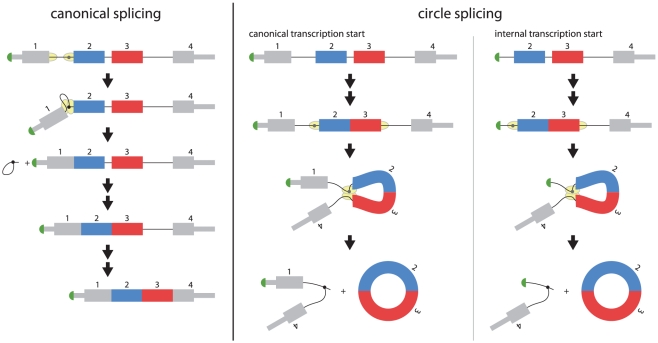
Figure 6. Models for generation of circular RNA.
At left: a schematic diagram of the canonical splicing process splicing out the first intron of the a pre-mRNA of a 4 exon gene, and subsequent removal of introns 2 and 3. Canonical splicing of exon 1 to exon 2 occurs when the splicing machinery catalyzes the formation of the intron lariat and the attack of the free 3′ OH of exon 1 on the 3′ splice site upstream of exon 2. This produces a lariat containing intron 1 and a pre-mRNA with exons 1 and 2 spliced together. At right: a model for the production of circular transcripts. If there is a canonical transcriptional start, and if intron excision does not proceed sequentially in time from the 5′ to 3′ direction of the pre-mRNA, non-canonical pairing of 3′ and 5′ splice sites could be generated. Since the sequences of each 5′ splice site of the pre-mRNA contain the same splicing signals, it is possible that the 3′ splice site upstream of exon 2 is paired with the 5′ splice site downstream of exon 3 and splicing proceeds as if this 5′ splice site were paired with the 3′ splice site upstream of exon 4. In this case, exon 3 would be spliced upstream of exon 2, creating a pre-mRNA intermediate comprised of these two exons and intron 2. Canonical splicing would be predicted to excise this intron, leaving a circular RNA composed of exons 2 and 3. Non-canonical transcription start, as suggested in [25], could produce an orphan 3′ splice site corresponding to the first transcribed exon. This splice site could be paired with a downstream 5′ splice site, generating a circular RNA. In both models, the excised intron would be linear and branched, and expected to be quickly degraded.