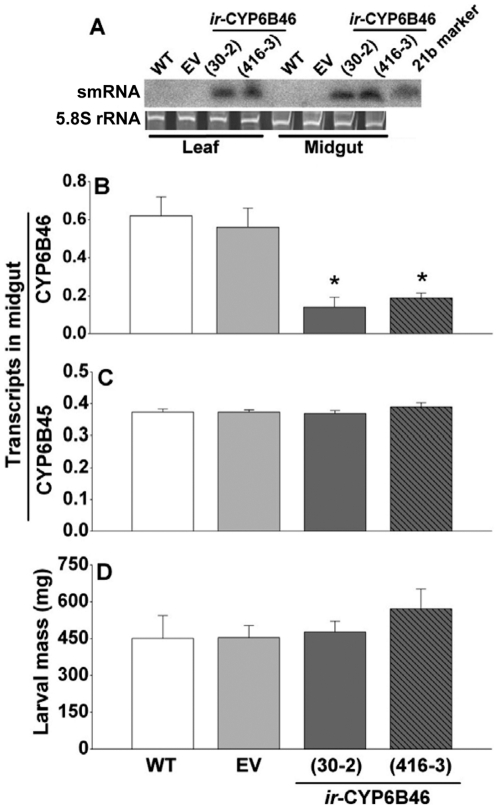
Figure 2. PMRi of M. sexta CYP6B46 using stably transformed N. attenuata plants.
(A) Northern hybridizations revealed the presence of small RNAs of CYP6B46 in the leaves of two independent lines of stably transformed plants, ir-CYP6B46 (30-2) and ir-CYP6B46 (416-3), and in the midguts of 4th instar larvae feeding on these plants. RNA samples from leaves of WT and EV (empty vector transformed stable line) plants and from the midguts of larvae feeding on WT and EV leaves were used as negative controls. Similar fluorescence intensity of the ethidium bromide stained 5.8 S rRNA bands reflected the equal loading of LMW RNA. Low molecular weight RNA from leaf or midgut loaded on the gel in each lane was a pool of three biological replicates. smRNA length of 21 b denoted by marker. Transcript abundance (relative to ubiquitin) of: (B) CYP6B46 (target gene) and closely related (C) CYP6B45 (off-target) in the midguts of 4th instar larvae. (D) Larval mass of 4th instar larvae feeding on WT, EV, ir-CYP6B46 (30-2) and ir-CYP6B46 (416-3) N. attenuata plants for 14 days. Asterisk indicates the significant differences as determined by one way ANOVAs (p≤0.05).