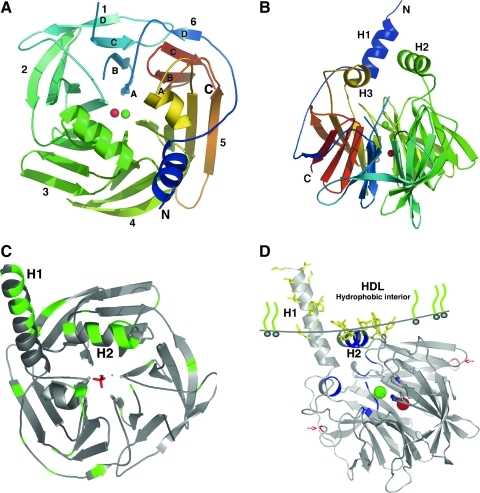
FIG. 3.
PON1 protein structure and its docking to HDL. (A) Top view of the six-bladed propeller-like structure of PON1. The top of the propeller shows the face that carries the loops linking the outer strand of each blade (strand D) with the inner strand (A) of the next blade. The N and C termini and the two calcium atoms (Ca1, green; Ca2, red) are located in the central tunnel of the propeller. 1–6, six blades of PON1 molecule. (B) One of the sides of the propeller. H1, H2, and H3 helices appear at the top of the propeller. Residues 1–15 (at the N-terminus) and residues 72–79 (in a surface loop between strands 1B and 1C) are invisible in this structural model [adapted from ref. (121) with permission]. (C) Tertiary structure of rePON1. The hydrophobic surfaces are shown to be exposed. N-terminal residues 7–18 are predicted to be helical and are thus modeled as part of H1, which is actually not displayed in the crystal structure. All the hydrophobic residues appear with an accessible surface area of 20 Å2. (D) PON1 is anchored to HDL via its hydrophobic side chains (shown in yellow). The line models the interface between the hydrophobic interior and the exterior aqueous portions of HDL, which is defined by the side chains of Tyr 185, Phe 186, Tyr190, Trp194, and Trp202 on helix H2 and the adjacent loops and Lys21 on helix H1, respectively. The hydrophobic side chains of the leucine and phenylalanine residues of H1 are located in the apolar region. The active site and the selectivity-determining residues are shown in blue. Glycosylation sites on Asn253 and Asn324 are shown in red. The high-resolution images were kindly supplied by Prof. Joel L. Sussman from the Weizmann Institute of Science, Israel. HDL, high-density lipoprotein.