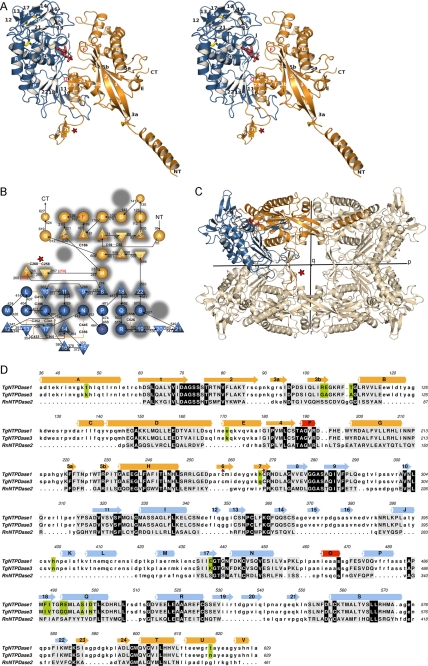
FIGURE 1.
Protein fold of TgNTPDase3. A, stereo view of a TgNTPDase3 monomer in complex with AMP (red). Domains I and II are colored in orange and blue, respectively. Strands are numbered 1–24, and helices are labeled A–V. Cysteines are depicted in yellow. B, topology diagram. Triangles, β-strands; circles, α-helices. The flanking strands 3 and 5 of the first β-sheet are interrupted by β-bulges. Red labeled helices and red marked β-sheet limitations correspond to the topology of the activated variant TgNTPDase3ΔCC. The topology of rat NTPDase2 (41) is underlaid in gray, highlighting the conservation, deletion, and addition of structural elements. C, orientation of the subunit domains in the tetramer. D, structure-based sequence alignment between TgNTPDase1 and -3 and RnNTPDase2 using RAPIDO (42, 52), with 328 aligned residues (root mean square deviation, 3.25 Å) and three defined rigid bodies of 159 residues (root mean square deviation (rigid bodies), 1.59 Å). Aligned residues are shown in capital letters and underlaid in gray. Residues conserved between TgNTPDases and RnNTPDase2 are underlaid in black, and positions that are different between TgNTPDase1 and -3 are highlighted in green. The red star in A–D highlights the activation loop with the disulfide bridge responsible for inactivity of the oxidized form.