FIGURE 1.
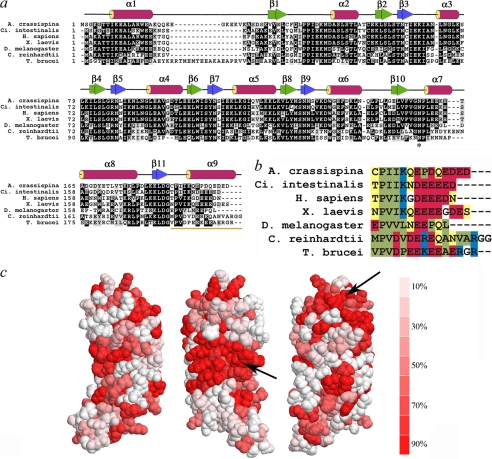
LC1 structural organization and conservation of C-terminal domain. a, ClustalW sequence alignment of LC1 orthologues from Anthocidaris crassispina (GenBank no. BAA24184), Ciona intestinalis (GenBank no. Q8T888), Homo sapiens (GenBank no. Q4LDG9), Xenopus laevis (GenBank no. Q28G94), Drosophila melanogaster (GenBank no. NP610483), C. reinhardtii (GenBank no. AAD41040), and T. brucei (GenBank no. XP828643). The secondary structure derived from the Chlamydomonas protein structure (Protein Data Bank code 1M9L) is indicated above the alignment. The asterisk indicates the conserved Asn residue that is mutated to Ser in the PCD patients. b, color-coded alignment of the C-terminal helix: polar, yellow; hydrophobic, green; acidic, red; basic, blue; glycine, white. c, the cross-species conservation is displayed on the LC1 surface: the graded colors represent the degree of conservation (identity) from red (≥90%) to white (≤10%). The arrows indicate major highly conserved regions of the molecular surface.