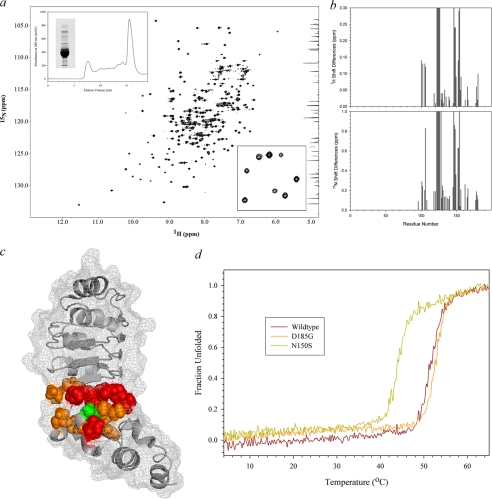
FIGURE 10.
Structural analysis of N150S PCD mutation. a, the entire 1H-15N HSQC spectrum of N150S LC1 is shown. The upper inset depicts the gel filtration and electrophoretic profiles of the mutant protein, and the lower inset shows an enlarged region of the spectrum illustrating that it is well resolved. Plots of the chemical shift changes for each residue in both dimensions are shown at right; missing/unidentified peaks were assigned values of 0.3 and 1.0 ppm for the 1H and 15N dimensions, respectively. b, the N150S mutation (green) and residues with chemical shifts that are >0.2 ppm different from wild type (orange) or missing/unidentified (red) are illustrated on the LC1 ribbon structure with surface mesh. Significant changes surround the mutation site and are propagated into the fourth and fifth LRRs. c, the thermal stability of wild-type, N150S, and D185G LC1 was assessed by circular dichroism spectroscopy in the range 4–65 °C. N150S LC1 was considerably destabilized and exhibited a Tm of 44 °C as compared with 51.5 and 52.5 °C for wild-type and D185G mutant LC1, respectively.