FIGURE 5.
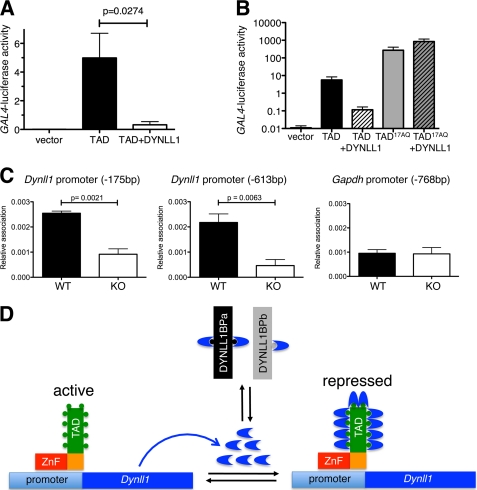
Autoregulation of DYNLL1 expression through feedback inhibition of the ASCIZ TAD. A, relative transcriptional activity of the isolated ASCIZ TAD tethered to the GAL4-luciferase reporter via the yeast Gal4-DNA binding domain in the absence or presence of overexpressed DYNLL1 in human U2OS cells; empty vector control is pCDNA3-Gal4DBD. Values are means ± S.E. of five independent experiments. B, similar experiments to panel A, also including the 17AQ-mutated ASCIZ TAD; n = 3 independent experiments. C, ChIP using the DYNLL1 antibody and quantitative real-time PCR analysis of the indicated regions of the Dynll1 or Gapdh promoters. Values are means ± S.E. of three to four different sets of WT and Asciz−/− littermate early-passage MEFs. D, proposed model of ASCIZ-dependent Dynll1 autoregulation. ASCIZ binds to the Dynll1 promoter via its ZnF domain and activates transcription via its TAD, resulting in expression of DYNLL1 protein (blue arcs). The equilibrium with other DYNLL1-binding proteins (DYNLL1BPa/b) determines how much DYNLL1 can bind to ASCIZ. At high levels of free DYNLL1, saturated binding to ten TQT motifs (circles) in the ASCIZ TAD represses Dynll1 transcription. Transcription is de-repressed when excess DYNLL1 is degraded or sequestered by increasing levels of DYNLL1-binding proteins.