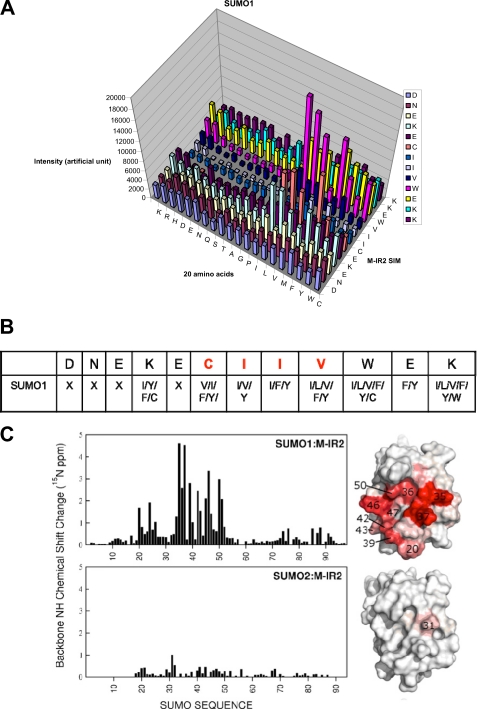
FIGURE 4.
Sequence requirements of the M-IR2 SIM. A, quantification of SUMO1 interactions with peptides from a peptide array based on the M-IR2 SIM (which binds in the antiparallel orientation). The SIM sequence is listed vertically with the N terminus at the bottom, and the 20 amino acid substitutions are listed horizontally. The intensity of each spot indicates the binding affinity toward SUMO, with taller bars indicating stronger interaction. B, summary of the amino acid preferences for binding SUMO1. X, no particular residue type is preferred. Amino acid residues are indicated by their one-letter codes. Red type indicates conserved residues. C, chemical shift perturbation analysis for the SUMO1-specific M-IR2 SIM with SUMO1. Changes in the backbone amide (15N-1H) chemical shift of SUMO are mapped on the surface representations of SUMO on the right and indicated by a white to red gradient to indicate zero to significant changes.