Background: RAD51C plays a key role in recombinational repair and genome stability.
Results: RAD51C deficiency leads to repair as well as a signaling defect in response to interstrand cross-links and double strand breaks.
Conclusion: RAD51C is critical for Fanconi anemia pathway of ICL repair and intra-S-phase checkpoint.
Significance: This study identifies distinct roles of RAD51C in repair and signaling and as a tumor suppressor.
Keywords: Cancer Biology, Checkpoint Control, DNA Damage Response, Genomic Instability, Radiation Biology, DNA Repair, Fanconi Anemia, Genome Stability, RAD51 Paralogs, Breast Cancer
Abstract
RAD51C, a RAD51 paralog, has been implicated in homologous recombination (HR), and germ line mutations in RAD51C are known to cause Fanconi anemia (FA)-like disorder and breast and ovarian cancers. The role of RAD51C in the FA pathway of DNA interstrand cross-link (ICL) repair and as a tumor suppressor is obscure. Here, we report that RAD51C deficiency leads to ICL sensitivity, chromatid-type errors, and G2/M accumulation, which are hallmarks of the FA phenotype. We find that RAD51C is dispensable for ICL unhooking and FANCD2 monoubiquitination but is essential for HR, confirming the downstream role of RAD51C in ICL repair. Furthermore, we demonstrate that RAD51C plays a vital role in the HR-mediated repair of DNA lesions associated with replication. Finally, we show that RAD51C participates in ICL and double strand break-induced DNA damage signaling and controls intra-S-phase checkpoint through CHK2 activation. Our analyses with pathological mutants of RAD51C that were identified in FA and breast and ovarian cancers reveal that RAD51C regulates HR and DNA damage signaling distinctly. Together, these results unravel the critical role of RAD51C in the FA pathway of ICL repair and as a tumor suppressor.
Introduction
Unrepaired or misrepaired chromosomal double strand breaks (DSBs)3 can cause gross chromosomal rearrangements that eventually can lead to tumorigenesis through inactivation of tumor suppressor genes or activation of oncogenes (1, 2). There are two major mechanisms of DSB repair, nonhomologous end joining and homologous recombination (HR) (3, 4). DSBs that are generated during the S- and G2-phase of the cell are preferentially repaired by sister chromatid recombination (SCR), an HR pathway that utilizes neighboring sister chromatid as a template (5, 6). Because the copied information is accurate, SCR is potentially an error-free pathway. HR also plays a critical role in the repair of daughter strand gaps (DSGs) that arise as a result of replication fork stalling and facilitates replication fork recovery (4, 7–9). Furthermore, in collaboration with nucleotide excision repair and translesion synthesis, HR is involved in the repair of DNA interstrand cross-links (ICLs) (10, 11). Thus, HR is important for the maintenance of genome integrity, and its dysfunction can lead to various genetic disorders and cancer (12–14).
The RAD51 recombinase plays a key role in the HR-mediated repair of DSBs, DSGs, and ICLs (15–17). The RAD51 paralog RAD51C is known to regulate HR in coordination with other RAD51 paralogs as follows: RAD51B, RAD51D, XRCC2, and XRCC3 (18). Biochemical studies show that RAD51C exists in two major complexes, RAD51B/RAD51C/RAD51D/XRCC2 (BCDX2) and the RAD51C/XRCC3 (CX3). Evidence from various studies shows that RAD51C participates in initial and late stages of HR (18). Chicken DT40 and hamster cells lacking RAD51C show reduction in RAD51 foci formation, decreased DSB repair by HR, and elevated chromosomal aberrations (18). RAD51C-deficient hamster cells display reduced repair of I-SceI-induced DSBs and increased frequency of long tract gene conversions (LTGC), suggesting the involvement of RAD51C in late stages of recombination (19). RAD51C has been shown to participate in DNA damage signaling by facilitating checkpoint effector kinase CHK2 phosphorylation (20).
Germ line mutations in BRCA1 and BRCA2 genes are known to cause hereditary breast and ovarian cancers (21, 22). BRCA1 and BRCA2 are required for HR-mediated DSB repair in mitotic and meiotic cells (23). These two proteins localize with RAD51 in a discrete nuclear foci in the S- and G2-phase of somatic cells. The interaction of BRCA1 and BRCA2 with RAD51 was further identified in a biochemical complex (24). The BRCA1-BRCA2-RAD51 complex localizes to the sites of stalled replication forks in response to hydroxyurea and ultraviolet irradiation, suggesting involvement of BRCA1 and BRCA2 in the HR-mediated repair of DNA lesions that arise as a result of replication fork stalling (24–26). More direct evidence for the BRCA1 and BRCA2 role in HR came from the fact that cells lacking BRCA1 or BRCA2 show reduced repair of DSBs induced by I-SceI expression (27, 28). RAD51 recombinase directly interacts with BRCA2 through BRC repeats that are present in the central part of BRCA2, and this association plays a pivotal role in the presynaptic assembly of RAD51 to the replication protein A-coated single-stranded DNA to facilitate HR (29). BRCA1 exists in multiple complexes to regulate various cellular processes, including checkpoint activation and HR to maintain genome integrity (30).
Fanconi anemia (FA) is a rare chromosome instability genetic disorder characterized by congenital abnormalities, progressive bone marrow failure, and susceptibility to cancer. 15 genes (FANCA–C, -D1, -D2, -E–G, -I and -J, and -L–P) have been identified to play a critical role in the FA pathway of ICL repair (18, 31, 32). Interestingly, three of the FA genes FANCD1, FANCJ, and FANCN are identified as breast cancer susceptibility (BRCA) genes BRCA2, BACH1, and PALB2, respectively (31, 32). Seven of the FA gene products (FANCA-C, -E-G, and -L) form an FA core complex to monoubiquitinate FANCD2 and FANCI (33, 34). FANCM protein with its binding partner FAAP24 have been shown to recruit FA core complexes to the sites of ICL lesions to monoubiquitinate FANCD2 and FANCI (35). The monoubiquitinated FANCD2 forms distinct nuclear foci that contain other key DNA damage response proteins like γ-H2AX, BRCA1, BRCA2, and RAD51 (36, 37). Failure of FANCD2 and FANCI monoubiquitination reduces ICL repair efficiency and HR (31, 32).
In a striking observation, germ line mutations in RAD51C have been identified to cause FA-like disorder and breast and ovarian cancers (38, 39). Biallelic mutation in RAD51C was identified in a family with a characteristic phenotype of FA. However, no hematological abnormalities or cancers were diagnosed in the children. Sequencing results confirmed the homozygous missense mutation (G773A, R258H) in RAD51C exon 5, and RAD51C has been assigned as FANCO, a 14th in the FA family genes (18, 38). In a parallel study, monoallelic mutations in RAD51C were identified to cause breast and ovarian cancers. A total of 14 mutations, which include base insertions, splice site mutations, and missense mutations were identified in RAD51C (18, 39). To understand the mechanistic insights into the role of RAD51C in ICL repair and its tumor suppressor function, here we have analyzed the RAD51C role in the FA pathway, DNA damage signaling, and repair. Our results clearly indicate that RAD51C is required for the HR step of ICL repair, a downstream role in the FA pathway. We show that RAD51C is critical for the repair of replication-associated DNA lesions. Strikingly, we identify a role for RAD51C in intra-S-phase checkpoint regulation. These results provide novel insights into the role of RAD51C in the FA pathway of ICL repair and as a tumor suppressor.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
The cell lines CL-V4B (RAD51C−/−) and V79B WT parental cells, which are Chinese hamster lung fibroblast cells and HeLa cells, were cultured as described previously (19). The stable cell lines of CL-V4B cells expressing WT and variant RAD51C from modified pcDNA3β vector were generated by transfection with Bio-Rad gene pulsar X cell (250 V and 950 microfarads), followed by selection in G418. The resistant colonies were pooled and expanded for functional assays.
DNA Constructs
The HR/SCR reporter and I-SceI expression vector design and construction were reported previously (40). The WT hRAD51C was PCR-amplified from the pcDNA3 hRAD51C vector and cloned into pcDNA3β vector using BamHI and XhoI sites. hRAD51C variants were generated by PCR-based mutagenesis and cloned in pcDNA3β vector using BamHI and XhoI sites and confirmed by sequencing. hRAD51C shRNA construct was generated using the reported RAD51C siRNA sequence (20) and cloned in pRS transient vector.
Immunoprecipitation, Western Blot, and Antibodies
Cells were harvested and lysed in RIPA buffer supplemented with complete protease and phosphatase inhibitor mixture (Roche Applied Science). For immunoprecipitation assays, cell lysates were incubated with anti-RAD51C polyclonal antibody (Santa Cruz Biotechnology) using protein A beads. Proteins were resolved on 10% SDS-PAGE and transferred onto PVDF membranes (Millipore). Membranes were blocked using 5% milk in TBST (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.1% Tween 20) and incubated with primary antibody for 1 h. The primary antibodies against RAD51C, RAD51, XRCC3, XRCC2, CHK1P (Ser-345), CHK2P (Thr-68), FANCD2, β-actin, and α-tubulin used for Western blot analysis were purchased from Santa Cruz Biotechnology. Membranes were incubated with secondary antibody and developed by chemiluminescence and captured using ChemiDoc (Fujifilm LAS 3000).
Chromosomal Aberration Assays
Cells were either mock-treated or treated with 10 ng/ml MMC for 24 h, followed by a 2-h incubation with 10 μg/ml colcemid. Later, cells were harvested and treated with hypotonic solution (0.5% KCl) for 20 min and fixed overnight at 4 °C in methanol/acetic acid (1:1). Cells were later dropped onto a chilled glass slide, and air-dried preparations were stained with 5% aqueous Giemsa. For each case, 50 metaphase plates were scored.
G2/M Accumulation Assays
Cells were incubated with 5 μm cisplatin or DMSO for 3 h and recovered in the fresh media for 36 h. Treated cells were trypsinized, and single cells were fixed overnight with 70% ethanol in PBS at −20 °C. After centrifugation, cells were incubated with 0.15 mg/ml RNase A (Fermentas) in PBS at 42 °C for 4 h and then incubated for 10 min with 50 μg/ml propidium iodide in the dark. A total of 1 × 104 cells were analyzed by Canto and caliber flow cytometer (BD Biosciences). Aggregates were gated out, and a percentage of cell with 4 n DNA content was calculated using FACSDiva Version 6.1.1 software (BD Biosciences).
Cell Survival Assays
Cells were seeded onto 24-well plates at a density of 5,000 cells per well in triplicate and were allowed to adhere for 4–6 h. Cells were mock-treated or treated continuously with MMC (Sigma), cisplatin (Sigma) for 3 h, MMS (Sigma) for 45 min, hydroxyurea (HU, Sigma) for 24 h, and camptothecin (CPT, Sigma) for 6 h. Cells were later recovered in the fresh media and incubated for 3–4 days. Later, 500 μl of 0.5 mg/ml MTT (Sigma) labeling reagent was added to each well, and the cultures were incubated for 5 h at 37 °C in dark. Later, the medium was removed, and 500 μl of DMSO was added to the cells; 100 μl of the resulting mixture was transferred to a 96-well plate and analyzed using microplate reader (VersaMax ROM Version 3.13). In a parallel experiment, cell viability was measured by counting using a hemocytometer. Percent growth was calculated as treated cells/untreated cell × 100.
HR Assays
HR/SCR assays were performed using 24 μg of I-SceI plasmid as described previously (19). In all experiments, the percentage of green GFP+ cells (whether I-SceI-induced or background frequency) was measured in triplicate samples, and I-SceI-transfected values were corrected for transfection efficiency. The spontaneous GFP+ frequency was subtracted from this number to obtain I-SceI-induced GFP+ frequency. The SCR events were quantified by BSDR+ cells as described previously (19).
Flow Cytometric Analysis of Cell Cycle
Cells were processed as discussed for G2/M accumulation studies. For the S-phase checkpoint experiment, cells were incubated with 3 μm CPT for 2 h. For bromodeoxyuridine (BrdU) incorporation assays, cells were treated with CPT for 2 h and recovered for up to 15 h followed by incubation with 60 μm BrdU (Sigma) for 20 min. Cells were fixed in 70% ethanol at 4 °C, and DNA was denatured using 2 n HCl and 0.5% Triton X-100 and later neutralized with 0.1 m sodium borate, pH 8.5. After that, cells were washed with PBS containing 0.5% Triton X-100 and 0.5% BSA and incubated with anti-BrdU mouse monoclonal antibody (BD Biosciences) for 2 h at room temperature. Later, cells were washed and incubated with anti-mouse secondary antibody conjugated with FITC (Sigma) for 1 h. After two washes in PBS, samples were incubated with RNase A and propidium iodide and analyzed by FACS. The aggregates and apoptotic cells were excluded, and cell cycle phases were analyzed by FACSDiva Version 6.1.1 software (BD Biosciences).
Comet Assays
The modified alkaline comet assay was performed using 20,000 cells as described previously (41). The MMC-treated cells were harvested at various time points to determine ICL formation and incision. Harvested cells were mixed with 150 μl of 0.5% low melting point agarose (Sigma) and spread on agarose-precoated frosted microscopic glass slides. Cells were lysed by placing slides in cold lysis solution (2.5 m sodium chloride, 100 mm EDTA, 10 mm Tris-HCl, pH 10, 1% Triton X-100, 10% DMSO) for 2 h. After lysis, slides were washed with PBS and incubated for 1 h in alkaline unwinding buffer (300 mm NaOH and 1 mm EDTA, pH >13) in the dark at 4 °C and later electrophoresed in the same buffer for 1 h at 25 V and 300 mA at 4 °C. Slides were neutralized in 0.4 m Tris-HCl, pH 7.5, and stained with ethidium bromide for analysis. Image of at least 50 cells per sample were captured by using a fluorescence microscope (Olympus IX 71), and tail moment was quantified using comet score.
Induction of DSBs in response to MMC treatment was evaluated by neutral comet assay as described previously (42). Briefly, cells were seeded in 12-well plates and treated with MMC continuously before harvesting at various time points. Harvested cells (20,000) were mixed with 500 μl of 1% low melting point agarose (Sigma) and spread on agarose-precoated frosted microscopic glass slides. Slides were placed in lysis solution (30 mm EDTA, 0.5% SDS, pH 8.3) and later incubated at 40 °C for 5 h. Following lysis, slides were washed with 90 mm Tris, 90 mm boric acid, 2 mm EDTA, and electrophoresed in the same buffer to 0.66 V/cm for 30 min. DNA was stained with 2 μg/ml propidium iodide for analysis. Images of individual cells were acquired using a fluorescence microscope.
RESULTS
RAD51C Is Crucial for Cross-link Repair
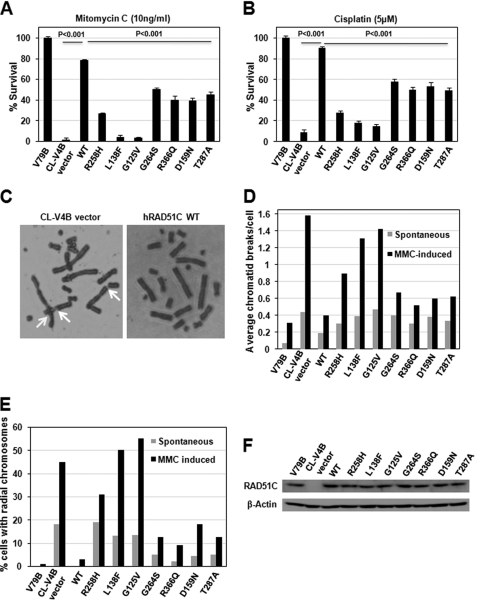
To understand the role of RAD51C in the FA pathway of ICL repair, here we have used Chinese hamster RAD51C-deficient cells. The residues that were found mutated in FA-like disorder, and breast and ovarian cancers are highly conserved in mouse and hamster cells (see supplemental Fig. S1A). The hallmarks of FA and BRCA cells are extreme sensitivity to ICL agents and chromosomal aberrations in the form of radial chromosomes and chromatid breaks. To understand the involvement of RAD51C in ICL repair, cell survival assays were carried out in the presence of various concentrations of ICL-inducing agents. CL-V4B empty vector cells showed extreme sensitivity to MMC and cisplatin, and stable expression of wild-type (WT) human RAD51C was able to rescue this phenotype (see supplemental Fig. S3, A and B). To understand the role of various RAD51C missense mutants that were identified in FA-like disorder and breast and ovarian cancers, we generated these mutants and stably transfected into CL-V4B cells as described under “Experimental Procedures.” At a fixed concentration of MMC and cisplatin, cells expressing RAD51C L138F and G125V missense mutants exhibited extreme sensitivity to ICL agents similar to that of empty vector cells, and four other mutants, G264S, R366Q, D159N, and T287A, as well as the R258H missense mutation that was identified in FA-like disorder showed moderate sensitivity, indicating the hypomorphic behavior of these mutants (Fig. 1, A and B). These results clearly suggest a critical role of RAD51C in ICL repair, and the pathological mutations in RAD51C compromise ICL repair. However, the expression levels of these mutants (Fig. 1F) and the growth pattern of CL-V4B cells expressing these RAD51C variants were not significantly different from the WTRAD51C-expressing cells (see supplemental Fig. S2A). To further confirm the involvement of RAD51C in the FA pathway, we analyzed metaphase spreads (Fig. 1C). Interestingly, absence of RAD51C or the cells expressing variant RAD51C exhibited a marked increase in the frequency of spontaneous as well as MMC-induced radial chromosomes as well as chromatid type breaks (Fig. 1, D and E), indicating a vital role of RAD51C in the FA and BRCA pathway of ICL and DSB repair. Strikingly, L138F and G125V mutations showed a remarkable (∼15-fold) increase in spontaneous as well as MMC-induced radial chromosomes and chromatid breaks, suggesting a critical role of these residues in the FA pathway of ICL repair. RAD51C G264S, R366Q, D159N, and T287A all showed moderate increase in chromosomal aberrations. Interestingly, the R258H missense mutation displayed a significant increase in spontaneous as well as MMC-induced chromosomal and chromatid-type aberrations, which is in agreement with sensitivity assays and suggests that R258H is a hypomorphic mutant (Fig. 1, D and E).
FIGURE 1.
RAD51C is essential for cross-link repair. RAD51C deficiency and pathological RAD51C mutants confers MMC (A) and cisplatin (B) sensitivity. C, metaphase spreads of CL-V4B empty vector or hRAD51C rescued cells exposed to 10 ng of MMC/ml. Arrows indicate chromatid breaks and radial chromosomes. Quantification of total chromatid breaks (D) and radial chromosomes (E) in CL-V4B vector and WTRAD51C rescued cells is shown. For each condition, 50 cells were analyzed. F, expression of WTRAD51C and its variants in CL-V4B cells. Actin was used as a loading control.
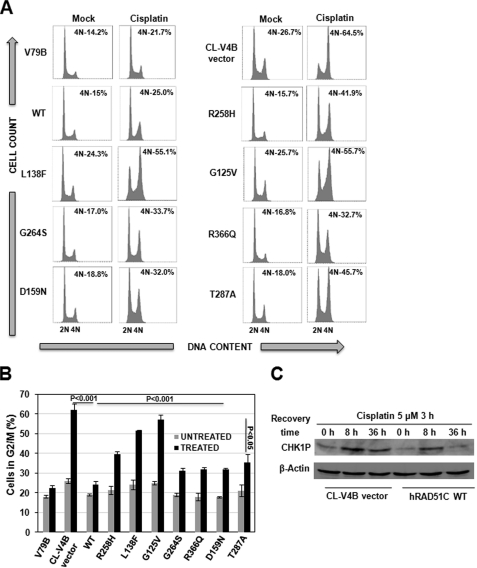
RAD51C Is Required for Normalization of Cell Cycle Response to ICL Agents
The FA cells show prolonged G2/M accumulation in response to ICLs. The transient G2/M arrest could allow cells to process ICLs for repair and reenter the cell cycle phases. To investigate the role of RAD51C in FA-BRCA pathway of ICL repair, we carried out cell cycle analysis of CL-V4B cells reconstituted with vector alone, WTRAD51C, and its variants after cisplatin treatment (Fig. 2, A and B). In contrast to parental V79B cells, CL-V4B empty vector cells displayed prolonged G2/M accumulation, and expression of WTRAD51C restored this abnormality, suggesting the RAD51C role in ICL repair. Expression of the R258H mutant suppressed G2/M accumulation to a moderate extent, indicating partial function of this mutant in ICL repair. In contrast, expression of L138F and G125V failed to suppress G2/M accumulation, although G264S, R366Q, D159N, and T287A showed a partial suppression compared with WTRAD51C. Similar observations have been made with MMC and melphalan (data not shown). This was further confirmed by the fact that CHK1 was activated for a longer duration in CL-V4B cells compared with WT cells (Fig. 2C), indicating the presence of an unprocessed pool of ICLs. These data again suggest that RAD51C is critical for ICL repair. To rule out the possibility that cell cycle kinetics is different in RAD51C mutant cells as compared with WT cells, we measured cell cycle distributions of the CL-V4B vector and WTRAD51C rescued cells at different time points after blocking the cells in G2/M-phase using nocodazole followed by release into fresh media and found no significant differences (see supplemental Fig. S2B).
FIGURE 2.
Germ line mutations in RAD51C leads to defective DNA damage response. A, cell cycle analysis of CL-V4B cells expressing WTRAD51C, RAD51C variants and vector control. Cells were treated with 5 μm cisplatin for 3 h and subsequently recovered for 36 h and analyzed by FACS to determine the G2/M cells. B, quantitative data for the experiment shown in A. Bar graph represents mean ± S.D from three independent experiments. C, CHK1 phosphorylation at indicated time points in CL-V4B vector and WTRAD51C-expressing cells.
RAD51C Is Dispensable for Incision of ICL and Monoubiquitination of FANCD2
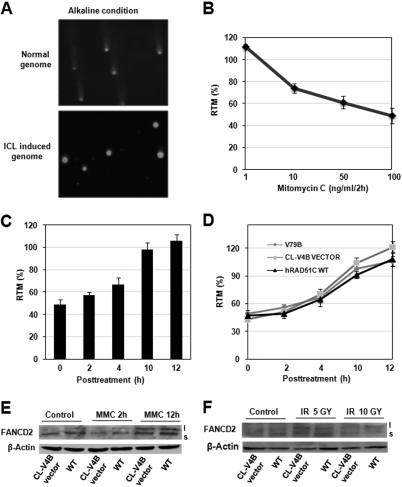
To test whether the sensitivity of RAD51C-deficient cells to the cross-linking agents is due to a defect in the incision step of cross-link repair, we measured the extent of DNA cross-linking by modified alkaline comet assay at single cell level (see “Experimental Procedures”) (Fig. 3A). Tail moment, a measure of relative electrophoretic mobility, decreased with increasing concentration of MMC for 2 h in V79B cells (Fig. 3B). Restoration of tail moment was observed with the recovery of treated V79B cells in fresh media in a time-dependent manner, indicating ICL unhooking and DSB formation (Fig. 3C). The efficiency of MMC-induced ICL unhooking in CL-V4B empty vector cells was compared with that of isogenic WTRAD51C rescued and parental V79B cells. The kinetics of incision in CL-V4B vector cells were indistinguishable from that of WTRAD51C complemented and V79B cells (Fig. 3D). This suggests that RAD51C is dispensable for incision of ICLs and acts subsequent to the formation of DSBs. One of the main steps in the FA pathway is the activation of FANCD2 by monoubiquitination, which is mediated by FA core complex. To investigate whether RAD51C is part of the FA core complex and plays a role in the activation of FANCD2, we measured FANCD2 monoubiquitination after MMC treatment. Interestingly, FANCD2 activation in CL-V4B vector cells was similar to WTRAD51C rescued cells (Fig. 3E). Similarly, ionizing radiation-induced FANCD2 monoubiquitination was not perturbed in CL-V4B empty vector cells compared with WTRAD51C complemented cells (Fig. 3F), suggesting that RAD51C acts downstream of FA core complex-mediated modification of FANCD2 and activation of the FA pathway.
FIGURE 3.
RAD51C is dispensable for ICL unhooking and activation of FA pathway. A, V7B untreated and treated cells with MMC (100 ng/ml, 2 h) were monitored for ICL induction and processing. B, dose-dependent decrease in relative tail moment in V79B cells compared with untreated cells. C, time-dependent restoration of relative tail moment in V79B cells after treatment with 100 ng/ml MMC for 2 h. D, quantification of relative tail moment in V79B, CL-V4B vector, and RAD51C rescued cells. For each cell type 50 comets were scored using comet score software. Analysis of FANCD2 monoubiquitination after MMC- (E) and ionizing radiation (F)-induced DNA damage. Gy, gray.
RAD51C Is Essential for Promoting Homology-directed I-SceI-induced DSB Repair
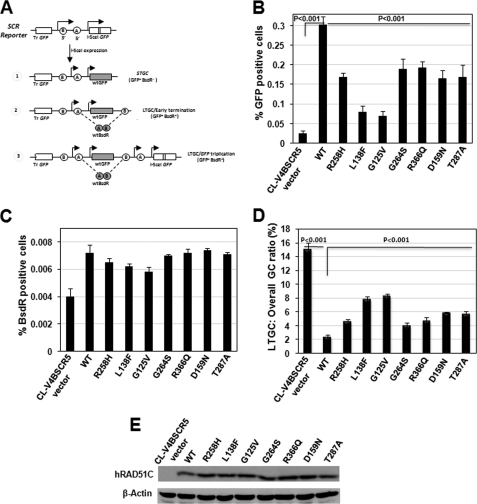
Cross-link repair essentially involves HR to complete the repair process. FA-BRCA pathway is known to be required for efficient HR, as depletion of BRCA1, BRCA2, FANCD2, FANCN, and FANCJ causes reduction in HR frequency, when measured by GFP-based HR reporter assay (32). We investigated whether pathological RAD51C mutants that were defective for ICL repair also display compromised HR. We used previously characterized CL-V4B SCR5 cells containing the HR/SCR reporter to study the HR events in response to a site-specific chromosomal DSB induced by I-SceI (19). Fig. 4A shows the description of the SCR reporter and the outcome of SCR events.
FIGURE 4.
RAD51C is required for promoting efficient HR. A, schematic diagram of the SCR reporter and the outcome of gene conversion events. I-SceI induced short tract gene conversion (STGC) that terminates prior to duplication of bsdR exon B (outcome 1), gene conversion that results in duplication of bsdR exon B but terminates prior to generating a third GFP copy, early terminating long tract gene conversion (LTGC/early termination) (outcome 2), and gene conversion that results in duplication of the bsdR cassette, generating a third GFP copy (LTGC/GFP triplication) (outcome 3). Wild-type GFP and BsdR mRNA is shaded. Tr GFP, truncated GFP. B, frequency of HR events measured as GFP+ cells of CL-V4B SCR reporter containing cells (CL-V4B SCR5) stably transfected with vector, WTRAD51C, and its variants. C, frequencies of I-SceI-induced BsdR+ colonies for the experiment shown in A. D, ratio of I-SceI-induced BsdR+/GFP+ frequencies (LTGC/overall GC, expressed as percentage) from the experiment whose results are shown in B and C. E, expression of RAD51C and various RAD51C mutant proteins or actin loading control for the experiment whose results are shown in B–D. The A is adapted with permission from Ref. 66.
Consistent with our previous observation, CL-V4B vector cells generated ∼0.02% I-SceI-induced GFP+ cells, and an ∼12-fold increase in the GFP+ cells was observed in the WTRAD51C rescued cells. However, expression of WTRAD51C resulted in only an ∼2-fold increase in the I-SceI-induced BsdR+ cells (Fig. 4, B and C). The corresponding ratio of LTGC/overall GC, a measure of the probability of a gene conversion event resolving as LTGC, was about 6-fold higher in control vector cells compared with WTRAD51C-expressing cells (Fig. 4D). These results are in agreement with a previous report (19) and suggest that RAD51C regulates overall GC and suppresses LTGC. In parallel experiments, expression of RAD51C L138F and G125V showed ∼4-fold reduction in the HR frequency. Cells expressing R258H, G264S, R366Q, D159N, and T287A displayed a partial HR defect compared with WT cells (Fig. 4B). The ratio of LTGC/overall GC was significantly increased (∼4-fold) in cells expressing RAD51C L138F and G125V, whereas R258H, G264S, R366Q, D159N, and T287A mutants showed a modest increase in LTGC events (Fig. 4D). The expression levels of various RAD51C mutants were similar to WT protein in CL-V4B SCR5 cells (Fig. 4E). These results clearly suggest that RAD51C residues that were found mutated in FA-like disorder and breast and ovarian cancers are essential for overall GC and in suppressing LTGC between sister chromatids.
RAD51C Is Critical for HR-mediated DSB and DSG Repair Induced by Clastogens
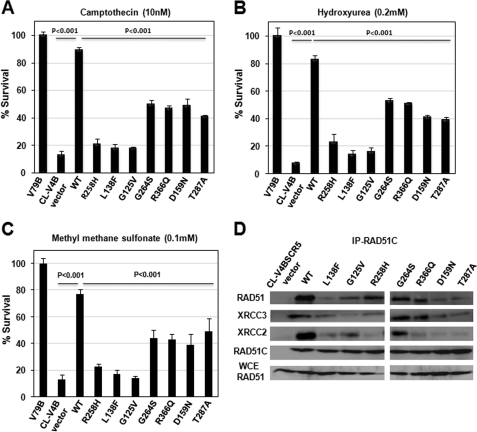
The HR measured using a GFP reporter reveals the repair of a single DSB induced by I-SceI in the entire genome. To understand the role of RAD51C in the repair of genome-wide DSBs and DSGs induced by clastogens such as topoisomerase inhibitor CPT, replication inhibitor HU, and MMS, we treated the cells with CPT, HU, and MMS and measured cell survival. Consistent with our HR data, CL-V4B control vector cells displayed extreme sensitivity to CPT, HU, and MMS with ∼10-fold reduction in survival, and this phenotype was rescued by WTRAD51C expression (see supplemental Fig. S3, C–E). RAD51C R258H, L138F, and G125V showed hypersensitivity similar to that of empty vector cells, suggesting the critical role of these residues in the RAD51C HR function. Cells expressing G264S, R366Q, D159N, and T287A displayed a partial reduction in cell survival compared with WTRAD51C rescued cells (Fig. 5, A–C). These results are in agreement with the data observed with HR reporter assay.
FIGURE 5.
RAD51C is essential for tolerance to genome-wide DSBs through RAD51-dependent HR. Sensitivity of CL-V4B vector, WTRAD51C, and its variant expressing cells to camptothecin (A), hydroxyurea (B), and MMS (C) at indicated concentrations. D, RAD51C interacts with RAD51 and forms complexes with XRCC3 and XRCC2. CL-V4B cells expressing WTRAD51C, its variants, and vector were treated with MMC (10 ng/ml, 12 h), and immunoprecipitation (IP) was carried out using anti-RAD51C antibodies. Immunoprecipitation products were analyzed using antibodies against RAD51, XRCC2, XRCC3, and RAD51C. Bottom lane, immunoblot with whole cell extracts (WCE) using an equivalent of 5% of total input extracts from all the samples, and all other lanes were loaded with 30% of the respective precipitates.
RAD51C has been shown to regulate RAD51, and its deficiency causes reduction in RAD51 foci formation at DNA damage sites; this could in turn lead to an HR defect. Our results with HR reporter and cell survival studies indicate the function of RAD51C in HR possibly through its interaction with RAD51. To test whether the compromised functions of various RAD51C mutants in HR are due to defective interaction with RAD51, we measured the efficiency of RAD51 interaction with RAD51C mutants as described under “Experimental Procedures.” We also measured the interaction of RAD51C mutants with XRCC3, the binding partner of RAD51C. Indeed, our data clearly show the physical association of RAD51C with RAD51 and XRCC3 (Fig. 5D). RAD51C L138F and G125V mutants showed a severe defect with ICL repair and HR assay. Consistent with these observations, interaction of RAD51C L138F and G125V mutants with RAD51 and XRCC3 was greatly diminished. The RAD51C R258H was partially functional with ICL repair and HR assay. In agreement with this, its interaction with both RAD51 and XRCC3 was partial. The G264S, R366Q, D159N, and T287A mutants were hypomorphic in our functional assays. Interestingly, the interaction of G264S and R366Q with RAD51 was partially reduced, but its association with XRCC3 was unperturbed. In the case of D159N and T287A RAD51C variants, the interaction was partial with XRCC3 but showed a greatly reduced association with RAD51. These results explain the possible reasons for the compromised functions of RAD51C mutants in ICL repair and HR.
Involvement of RAD51C in MMC-induced DNA Damage Response
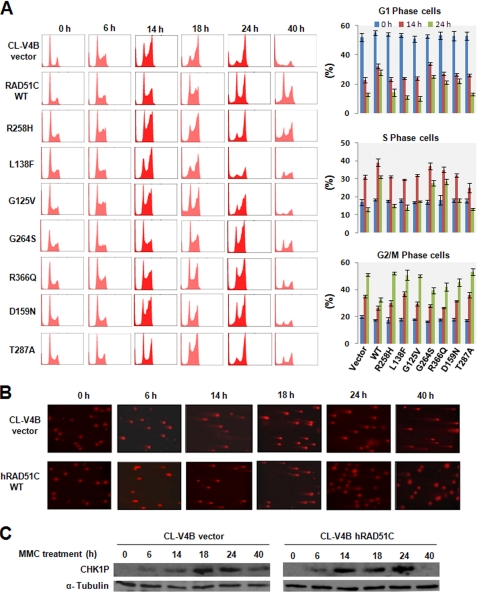
To understand whether RAD51C participates in ICL-induced DNA damage response, we treated CL-V4B empty vector and WTRAD51C rescued cells to MMC and analyzed the cell cycle profile at various time points (see “Experimental Procedures”). The cell cycle profile of the CL-V4B vector and RAD51C rescued cells were very similar after 6 h of MMC treatment. Interestingly, at 14 h, CL-V4B vector cells showed a predominant population in G2/M-phase (Fig. 6A). In contrast, WTRAD51C rescued cells displayed significant population in S-phase. Strikingly, at this time point, accumulation of DSBs and CHK1 activation was similar in CLV-4B vector and RAD51C rescued cells (Fig. 6, B and C). Notably, progression of CL-V4B vector cells into G2/M-phase was robust by 24 h despite the presence of abundant DSBs (Fig. 6, A and B). In contrast to control cells, RAD51C rescued cells showed retarded progression and reduced DSBs (Fig. 6, A and B), suggesting the activation of checkpoint and efficient repair of ICLs. However, WTRAD51C rescued cells displayed near normal cell cycle profile by 40 h and inactivation of CHK1, further suggesting the completion of ICL repair in these cells. Strikingly, the vector cells showed accumulated DSBs even at 40 h with a prolonged activation of CHK1 and underwent significant cell death from G2/M arrest (Fig. 6, A–C). The unperturbed progression of CL-V4B cells into G2/M-phase despite MMC-induced ICL lesions indicates the possible role of RAD51C in S-phase checkpoint.
FIGURE 6.
RAD51C is required for MMC-induced S-phase checkpoint control and repair. A, indicated cells were treated with 50 ng/ml MMC continuously, harvested at indicated time points, and analyzed by flow cytometry to determine the percentage of cells at different phases of cell cycle. A representative experiment is shown. The bar graph represents mean ± S.D. B, representative neutral comet images for the CL-V4B vector and WTRAD51C complemented cells at various time points after MMC treatment. C, CL-V4B vector and WTRAD51C rescued cells collected after MMC treatment were lysed and analyzed for CHK1 activation.
Analyses of RAD51C variants showed that cells expressing R258H, G125V, and D159N displayed cell cycle progression faster than WTRAD51C-expressing cells but slower than vector cells at 14 h. Accumulation of DSBs at this time point was nearly identical in these mutants (see supplemental Fig. S4). Analyses of cell cycle progression at 24 h revealed that these mutants accumulated in G2/M-phase and showed partial release from G2/M arrest by 40 h (Fig. 6A), suggesting a partial defect in these mutants. Interestingly, the cell cycle profile of L138F and T287A showed a phenotype similar to empty vector cells at the indicated time points, implying the severe defect of these mutants in DNA damage response (Fig. 6A). The RAD51C variants G264S and R366Q showed near normal cell cycle profile similar to WTRAD51C rescued cells at 14 h. However, by 24 h, these mutants showed more cells in G2/M and at 40 h displayed a partial release from G2/M (Fig. 6A), suggesting the hypomorphic behavior of these mutants in ICL-induced DNA damage response.
RAD51C Plays a Critical Role in the Intra-S-phase Checkpoint
Our results with defective S-phase checkpoint in RAD51C-deficient CL-V4B cells to MMC-induced DNA damage prompted us to investigate whether RAD51C controls S-phase checkpoint in response to CPT. To investigate this, we treated CL-V4B empty vector and WTRAD51C rescued cells with CPT and analyzed the cell cycle progression as described under “Experimental Procedures.” Both these cells showed progression with a similar kinetics for up to 2 h of recovery following 3 h of CPT treatment. Interestingly, in the later hours, WT rescued cells showed delayed cell cycle progression with more of an S-phase population as compared with vector cells that displayed early entry into G2/M-phase (see supplemental Fig. S5). The accumulation of WTRAD51C rescued cells in S-phase suggests the intact S-phase checkpoint in these cells. In contrast, control vector cells failed to arrest in S-phase in response to CPT-induced DNA damage, indicating a role of RAD51C in S-phase checkpoint response. Our analyses with various RAD51C mutants showed a similar phenotype as that was found with MMC-induced DNA damage (see supplemental Fig. S5).
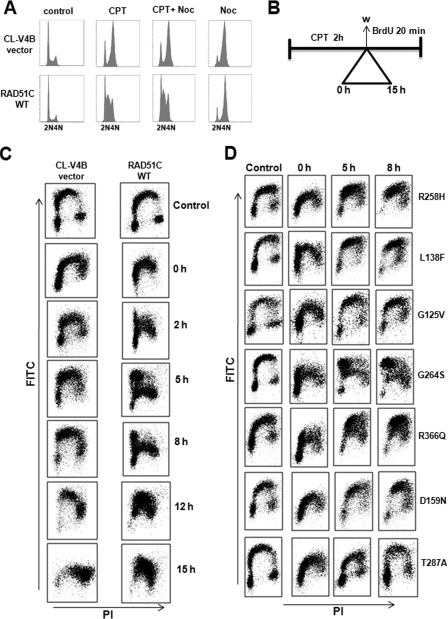
To further validate our observation that RAD51C regulates S-phase checkpoint, cells were trapped in G2/M-phase using the microtubule inhibitor nocodazole. Both WTRAD51C and vector-transfected CL-V4B cells accumulated in G2/M-phase after nocodazole treatment for 15 h. Cotreatment of cells with nocodazole and CPT for the indicated time intervals showed that WT cells were not affected by nocodazole treatment because of intact S-phase checkpoint function (Fig. 7A). In contrast, control vector cells showed progression into G2/M-phase after treatment with CPT and arrested in G2/M-phase when cotreated with nocodazole. These results point to the novel role of RAD51C in DNA damage-induced S-phase checkpoint regulation.
FIGURE 7.
RAD51C is critical for the intra-S-phase check point after replication stress. A, CL-V4B vector cells and CL-V4B cells stably transfected with WTRAD51C were either untreated or treated with CPT (1.5 μm for 3 h), nocodazole (NOC) (150 ng/ml), or both, and cells were harvested after 15 h and subjected to flow cytometry. B, experimental protocol for measuring intra-S-phase checkpoint. C, BrdU+ cells were monitored for the CL-V4B cell transfected with vector or WT RAD51C after treatment with 3 μm CPT for 2 h and pulse-labeled with BrdU before harvesting the cells at the indicated time and processed for analysis as described under “Experimental Procedures.” Incorporation of BrdU (y axis) with respect to total DNA content (x axis) was analyzed by flow cytometry. D, BrdU+ incorporation for RAD51C variants was monitored after treatment with 3 μm CPT for 2 h and pulse-labeled with BrdU before harvesting the cells at the indicated time and processed for analysis as described under “Experimental Procedures.”
The DNA damage response regulator ATM and FA-BRCA pathway proteins FANCD1 (BRCA2), FANCD2, and FANCN (PALB2) have been shown to regulate the intra-S-phase checkpoint. Mutation in any of these genes shows radio-resistant DNA synthesis (43). Our findings of RAD51C having a role in the S-phase checkpoint prompted us to investigate whether RAD51C also regulates intra-S-phase checkpoint similar to FA-BRCA pathway proteins. We examined whether RAD51C can suppress DNA synthesis in the presence of DSBs, which is the hallmark of intra-S-phase checkpoint. To test this hypothesis, we carried out BrdU incorporation after treatment with CPT as described under “Experimental Procedures” (Fig. 7B). BrdU pulse labeling failed to indicate a difference in DNA synthesis in WT versus control vector cells in untreated samples. Strikingly, DNA synthesis was strongly inhibited at 0 h of recovery in WT rescued cells, and some populations of CL-V4B vector cells underwent DNA replication. From 2 to 8 h of recovery, CL-V4B vector cells resumed DNA synthesis in early to mid S-phase, whereas DNA synthesis in WT RAD51C cells was limited only to early S-phase. At 12 h of recovery, the majority of CL-V4B cells entered late S-phase followed by G2-phase at 15 h, whereas WTRAD51C cells were synthesizing DNA in mid to late S-phase during the same recovery hour. These results demonstrate that RAD51C regulates intra-S-phase checkpoint (Fig. 7C).
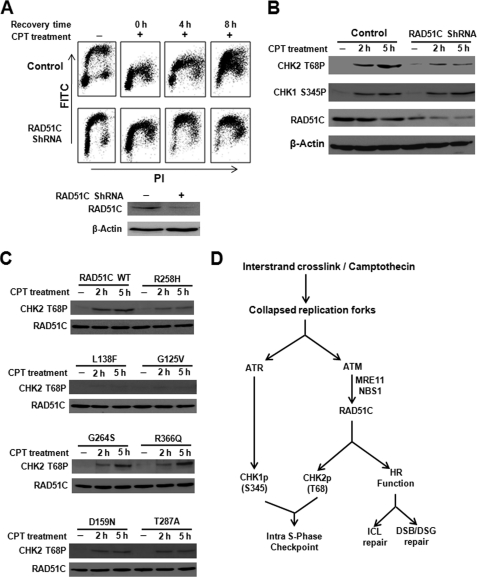
Analysis of RAD51C mutants revealed that R258H, L138F, G125V, D159N, and T287A caused unperturbed DNA replication similar to empty vector cells. In contrast, R366Q showed a partial suppression of DNA synthesis, indicating the hypomorphic behavior of this mutant in the intra-S-phase checkpoint control. Strikingly, although G264S mutant was partially defective in ICL repair and HR, it suppressed DNA synthesis and exhibited a phenotype similar to WT rescued cells in the presence of DSBs (Fig. 7D). Similar observations were made in RAD51C-depleted HeLa cells, and DNA synthesis in control cells was strongly inhibited at 4 and 8 h of recovery after CPT treatment. A significant population of RAD51C shRNA-treated cells underwent robust DNA replication from early to mid and late S-phase (Fig. 8A). Fig. 8B shows that down-regulation of RAD51C leads to reduced activation of CHK2 after CPT treatment, although CHK1 activation was unaffected. To test whether pathological mutants of RAD51C affect CHK2 activation, we transfected HeLa cells with WT and variants of RAD51C and examined CHK2 activation (Fig. 8C). CHK2 phosphorylation was intact in cells expressing WTRAD51C but overexpression of R258H, D159N, and T287A reduced the activation of CHK2. Notably, L138F and G125V completely abrogated CHK2 activation. In contrast, G264S and R366Q variants showed near normal CHK2 activation similar to WT RAD51C-expressing cells. Together, these data suggest that RAD51C regulates the intra-S-phase checkpoint in response to CPT-induced DSBs by activating CHK2. Based on our detailed analyses, we propose a model (Fig. 8D) to show the dual role of RAD51C in genome maintenance by promoting efficient repair and DNA damage signaling by ATM-mediated activation of CHK2.
FIGURE 8.
RAD51C regulates intra-S-phase checkpoint through CHK2 activation in response to replication-associated DSBs. A, knocking down RAD51C in HeLa cells leads to loss of intra-S-phase checkpoint control after CPT (1 μm for 2 h) treatment as measured by BrdU incorporation. B, RAD51C depletion in HeLa cells decreases CHK2 phosphorylation after CPT treatment. Control as well as cells transfected with RAD51C shRNA were treated with 1 μm CPT for the indicated time. C, overexpression of various RAD51C variants affect CHK2 activation after CPT treatment. HeLa cells were transfected with 1 μg of WTRAD51C and RAD51C variants treated with 1 μm CPT for the indicated time. D, model to explain RAD51C-regulated HR and intra-S-phase checkpoint in response to ICL and replicative stress.
DISCUSSION
In this study, we demonstrate that RAD51C regulates the FA pathway of ICL repair. We show that RAD51C is dispensable for ICL unhooking as well as activation of the FA pathway. However, our study shows that RAD51C participates in the HR step of ICL repair. In addition, our results indicate that RAD51C is essential for the repair of genome-wide DSBs and DSGs that arise during replication. Although RAD51C has been implicated in DNA damage signaling by regulating CHK2 phosphorylation (20), its precise role in DNA damage-induced cell cycle checkpoint is unknown. In this study, for the first time we unequivocally demonstrate that RAD51C regulates intra-S-phase checkpoint through CHK2 activation. Data presented here with RAD51C variants clearly identify distinct roles of RAD51C in DNA damage signaling and repair by HR.
The hallmarks of FA cells are extreme sensitivity to ICL agents and chromatid-type errors in the form of radial chromosomes and chromatid breaks (44). Indeed, RAD51C-deficient hamster cells displayed remarkable sensitivity to MMC and cisplatin. In addition, ICL-induced radial chromosomes as well as chromatid breaks were abundant in RAD51C-deficient cells, suggesting that these cells are defective for ICL repair. FA cells also display checkpoint activation and accumulate in G2/M as a response to unrepaired ICLs. In fact, RAD51C-deficient cells showed extended ATR-dependent CHK1 phosphorylation and prolonged accumulation at G2/M. These results confirm the participation of RAD51C in the FA pathway of ICL repair.
The repair of ICLs has been thought to begin with unhooking of lesions by specific nucleases. The heterodimeric nucleases MUS81-EME1 and XPF-ERCC1 have been implicated in ICL unhooking. Recently, the FAN1 and SLX1-SLX4 complex also has been identified as nucleases involved in the processing of ICLs, and biallelic mutation in SLX4 has been recently identified to cause FA (31, 32). We show here that RAD51C is dispensable for processing of ICLs but plays a role downstream in the ICL repair. Activation of the FA pathway by FANCD2 monoubiquitination is a key event in ICL repair, and mutations in components of the FA core complex are known to affect FANCD2 monoubiquitination (32). Notably, mouse knock-out of RAD51C leads to early embryonic lethality (45), and our data with the RAD51C R258H that was identified to cause FA-like disorder provides evidence that RAD51C R258H mutation is hypomorphic. FANCD2 monoubiquitination was not perturbed with cells expressing RAD51C R258H (38). Because this was a hypomorphic mutant, we argued in a review article (18) and investigated here whether RAD51C deficiency leads to a defect in the activation of FANCD2. Interestingly, MMC-induced levels of FANCD2 monoubiquitination were unaltered in RAD51C-deficient cells compared with WTRAD51C rescued cells. This again suggests that RAD51C plays a downstream role, possibly at the HR step in the ICL repair.
We have previously reported that RAD51C regulates overall GC and suppress LTGC between sister chromatids (19). However, whether short tract gene conversions and LTGC represent two different pathways and whether RAD51C regulates these pathways distinctly is currently unknown. In our studies, RAD51C L138F and G125V mutants showed greatly reduced overall GC, and the remaining mutants showed partial reduction. Notably, the relative decrease in overall GC in these mutants was associated with a concomitant increase in LTGC. Thus, it remains unclear how RAD51C differentially regulate short tract gene conversions and LTGC. HR plays a major role in the repair of DNA lesions associated with replication and in the recovery of stalled/collapsed replication forks (7, 9, 17, 46). RAD51C-deficient cells were hypersensitive to genotoxic agents that induce genome-wide DSBs and DSGs, indicating the critical role of RAD51C in HR function. This was further supported by the fact that RAD51C-deficient cells were defective in cell cycle progression upon HU-induced replication stress, indicating the crucial role of RAD51C in HR-mediated replication fork recovery (data not shown). Indeed, RAD51C has been shown to accumulate at HU-induced stalled replication forks (20). The HR-defective phenotype observed with various RAD51C mutants corroborates with the ICL sensitivity data, suggesting that the role of RAD51C in ICL repair is attributed to the HR function. Together, these observations strongly indicate that RAD51C HR function is crucial for the repair of ICLs, DSBs, and DSGs. However, mechanistically it is not known how RAD51C is executing the repair of these lesions by HR. Notably, RAD51C has been shown to interact with RAD18 and execute DNA damage repair by HR (47). Conceivably, RAD51C may exist in different repair complexes to execute the repair of various types of lesions by HR. RAD51 is a key player in HR, and RAD51C has been shown to interact with RAD51 (48, 49). In our studies, RAD51C mutants showed defective interaction with RAD51, and this may explain the compromised functions of these mutants for HR-mediated repair of ICLs, DSBs, and DSGs.
RAD51C has been shown to be important for efficient DNA damage signaling by regulating CHK2 phosphorylation in response to ionizing radiation (20), suggesting that RAD51C could act as a mediator or amplifier in ATM-dependent DNA damage signaling. However, the molecular mechanism by which RAD51C controls this process is unclear. Interestingly, we find that RAD51C-deficient cells failed to arrest in S-phase in response to MMC- and CPT-induced DSBs but arrest in G2/M. In contrast, WTRAD51C rescued cells arrested in S-phase, suggesting that RAD51C may have a role in intra-S-phase checkpoint function. The hallmark of intra-S-phase checkpoint defect is damage-resistant DNA synthesis (50). Indeed, BrdU incorporation measured after CPT treatment was unperturbed in RAD51C-deficient cells compared with WTRAD51C-expressing cells, indicating that RAD51C regulates intra-S-phase checkpoint. Notably, FANCD2 also has been shown to control intra-S-phase checkpoint in an ATM- and NBS1-dependent fashion (51). It will be interesting to study whether there is any cross-talk between RAD51C and FANCD2 in the regulation of intra-S-phase checkpoint. Previous studies have shown that MRE11 and NBS1 regulate intra-S-phase checkpoint in an ATM- and CHK2-dependent fashion (52, 53). Interestingly, NBS1 depletion was reported to reduce RAD51C foci formation at the sites of DSBs (20). Our results with CPT-induced DSBs clearly show reduced CHK2 phosphorylation in RAD51C-depleted cells, suggesting that RAD51C regulated intra-S-phase checkpoint in response to replication-associated DSBs is indeed through ATM (Fig. 8D). RAD51C possess conserved SQ/TQ motifs that are targets for ATM/ATR phosphorylation. However, it is unclear whether these motifs are substrates for ATM/ATR kinase and whether these phosphorylations are required for DNA damage responses. Our data with RAD51C G264S and R366Q mutants show that these mutants are normal for intra-S-phase checkpoint function. Nonetheless, these mutants were defective for repair by HR, implying the distinct roles of RAD51C in DNA damage signaling and repair. It is likely that RAD51C containing different complexes may be regulating DNA damage signaling and repair independently. Notably, RAD51C G264S and R366Q showed near normal interaction with XRCC3, suggesting that the CX3 complex may be involved in the regulation of checkpoint function. Indeed, XRCC3 depletion also was found to affect CHK2 phosphorylation (20). The CX3 complex also has been implicated in late stages of HR (54). However, mechanistically how the CX3 complex controls these independent functions requires further investigation.
BRCA1 tumor suppressor is known to have multiple functions, including its roles in DNA damage signaling, HR, and genome stability, and these functions have been implicated in tumor suppressor function (55). BRCA1 exists in multiple complexes, and these complexes are known to have independent as well as overlapping functions in genome maintenance (55). Our studies provide evidence for the multiple functions of RAD51C in DNA damage response and repair by HR. It is possible that RAD51C exists in complexes that are required for mediating checkpoint functions in response to DNA damage. RAD51C may play an independent role in HR through a different complex containing HR factors. Further studies are required to understand whether RAD51C is part of a multiple complex similar to BRCA1 in executing various cellular functions. BRCA1 has been implicated as a scaffold protein providing a platform for assembly of various cellular proteins that regulate DNA damage signaling and repair. Notably, RAD51C possesses nuclear localization signal and has been shown to mobilize RAD51 from cytoplasm to nucleus in response to DNA damage (56). The RAD51C R366Q is a mutation in the NLS motif and shows partial loss of function in our assays. Indeed, deletion of RAD51C NLS sequences is known to cause hypersensitivity to MMC (57). Despite RAD51C being a small protein lacking defined motifs, similar to BRCA1, RAD51C may also provide scaffolding function to mediate HR function as well as signaling.
Data presented here clearly show that pathological RAD51C mutants are defective in interaction with RAD51. Although BRCA2 has been shown to play a mediator role in RAD51 loading onto the sites of DNA damage, RAD51C might also have a similar function in RAD51 recruitment to the sites of DNA lesion. Consistent with this hypothesis, BRCA2-dependent and -independent RAD51 foci formation has been reported (58). Furthermore, nuclear RAD51 levels in BRCA2-deficient cells were further reduced when RAD51C was depleted (56). These data support the critical role of RAD51C in RAD51 regulation. However, the underlying mechanism by which BRCA2 and RAD51C regulate RAD51 loading independently at the sites of DNA lesion is currently elusive.
The mechanism by which RAD51C controls distinct functions of repair and checkpoint response is unclear. As we discussed previously, RAD51C lacks any defined motifs except for conserved Walker A (GXXXXGKT) and Walker B (LLIVD) motifs, as required for ATPase function found in RecA/RAD51 family proteins (see supplemental Fig. S1B). Our detailed analyses of pathological mutants of RAD51C indicate that Gly-125 and Leu-138 are critical for repair as well as signaling functions. Interestingly, Gly-125 is a conserved residue in the Walker A motif, suggesting that RAD51C ATPase function is important for repair and signaling. The RAD51C Leu-138 is a conserved residue in RAD51 and is located next to the Walker A motif. The phenotype of the L138F mutant was very similar to G125V. It is likely that this residue may have a role in RAD51C ATPase function. The RAD51C R258H is hypomorphic in repair and checkpoint functions of RAD51C. The N-terminal domain of RAD51 is important for DNA binding activity. The RAD51C R258H has been speculated to affect the N-terminal and ATPase domains of RAD51C (38). RAD51C G264S, R366Q, D159N, and T287A mutants display partially reduced repair function. Notably, G264S was completely normal for signaling, whereas R366Q showed a near normal function for signaling. The Gly-264 and Thr-287 residues are not conserved in RAD51, and currently the role of these residues in RAD51C repair and checkpoint functions is unclear. Asp-159 is a conserved residue in RAD51, but the clear role of this residue in RAD51 is unknown. The RAD51C Arg-366 is located in the NLS motif of RAD51C (57) and appears to have an important role in RAD51C mobilization to the nucleus.
Monoallelic mutations in BRCA1, BRCA2 (FANCD1), PALB2 (FANCN), and BACH1 (FANCJ) are known to cause hereditary breast and ovarian cancers. Biallelic inactivation of BRCA2, PALB2, and BACH1 leads to FA (59). Alterations in any of these genes show extreme sensitivity to ICL agents as these agents block replication abruptly. Defective processing of such lesions is thought to cause structural alterations in the form of chromatid breaks, tri- and quadriradial chromosomes, as well as gross chromosomal rearrangements. Indeed, FA and BRCA cells show accumulation of such types of structural alterations in the chromosomes (60). The major functions of FA and BRCA proteins are in dealing with DNA lesions that arise during DNA replication through HR, implying the crucial role of HR in maintaining genome integrity and tumor suppression (61). Strikingly, RAD51C-deficient cells also displayed chromatid-type errors, providing evidence that RAD51C is a novel component in the FA and BRCA pathway of DNA repair. BRCA pathway also plays a critical role in the repair of genome-wide DSBs and DSGs that arise during replication (7, 9, 26). The BRCA proteins BRCA1, BRCA2, and PALB2 have been shown to regulate intra-S-phase checkpoint (62–65). The cell cycle checkpoint and the HR functions controlled by BRCA proteins are essential for repairing DSBs and DSGs to maintain genome stability and prevent tumorigenesis. Similar to BRCA proteins, this study provides evidence for the role of RAD51C in tumor suppression through HR and DNA damage signaling functions. However, further studies are required to understand how RAD51C mechanistically regulates various cellular functions.
Supplementary Material
Acknowledgments
We are grateful to Drs. Kum Kum Khanna, Sandeep Burma, and K. Muniyappa for their critical comments on the manuscript. We thank Drs. K. Muniyappa and D. N. Rao for their support and encouragement. We thank Dr. Omana Joy, Kavya Ananthaswamy and Pooja Pai at IISc FACS facility for their help on cell cycle analyses.
This work was supported in part by the Indian Institute of Science, Department of Biotechnology, Council of Scientific and Industrial Research, and Department of Science and Technology.

This article contains supplemental Figs. 1–5.
- DSB
- double strand break
- HR
- homologous recombination
- FA
- Fanconi anemia
- ICL
- interstrand cross-link
- DSG
- daughter strand gap
- HU
- hydroxyurea
- CPT
- camptothecin
- MMC
- mitomycin C
- MMS
- methyl methane sulfonate
- SCR
- sister chromatid recombination
- LTGC
- long tract gene conversion
- ATM
- ataxia telangiectasia mutated
- ATR
- ATM and Rad3-related protein
- GC
- gene conversion.
REFERENCES
- 1. Aguilera A., Gómez-González B. (2008) Genome instability. A mechanistic view of its causes and consequences. Nat. Rev. Genet. 9, 204–217 [DOI] [PubMed] [Google Scholar]
- 2. Negrini S., Gorgoulis V. G., Halazonetis T. D. (2010) Genomic instability. An evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 11, 220–228 [DOI] [PubMed] [Google Scholar]
- 3. Moynahan M. E., Jasin M. (2010) Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 11, 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ciccia A., Elledge S. J. (2010) The DNA damage response. Making it safe to play with knives. Mol. Cell 40, 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johnson R. D., Jasin M. (2000) Sister chromatid gene conversion is a prominent double strand break repair pathway in mammalian cells. EMBO J. 19, 3398–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takata M., Sasaki M. S., Sonoda E., Morrison C., Hashimoto M., Utsumi H., Yamaguchi-Iwai Y., Shinohara A., Takeda S. (1998) Homologous recombination and nonhomologous end-joining pathways of DNA double strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17, 5497–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nagaraju G., Scully R. (2007) Minding the gap. The underground functions of BRCA1 and BRCA2 at stalled replication forks. DNA Repair 6, 1018–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Branzei D., Foiani M. (2010) Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11, 208–219 [DOI] [PubMed] [Google Scholar]
- 9. Budzowska M., Kanaar R. (2009) Mechanisms of dealing with DNA damage-induced replication problems. Cell Biochem. Biophys. 53, 17–31 [DOI] [PubMed] [Google Scholar]
- 10. Räschle M., Knipscheer P., Knipsheer P., Enoiu M., Angelov T., Sun J., Griffith J. D., Ellenberger T. E., Schärer O. D., Walter J. C. (2008) Mechanism of replication-coupled DNA interstrand cross-link repair. Cell 134, 969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Muniandy P. A., Liu J., Majumdar A., Liu S. T., Seidman M. M. (2010) DNA interstrand cross-link repair in mammalian cells, step by step. Crit. Rev. Biochem. Mol. Biol. 45, 23–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McKinnon P. J., Caldecott K. W. (2007) DNA strand break repair and human genetic disease. Annu. Rev. Genomics Hum. Genet. 8, 37–55 [DOI] [PubMed] [Google Scholar]
- 13. O'Driscoll M., Jeggo P. A. (2006) The role of double strand break repair. Insights from human genetics. Nat. Rev. Genet. 7, 45–54 [DOI] [PubMed] [Google Scholar]
- 14. Venkitaraman A. R. (2009) Linking the cellular functions of BRCA genes to cancer pathogenesis and treatment. Annu. Rev. Pathol. 4, 461–487 [DOI] [PubMed] [Google Scholar]
- 15. San Filippo J., Sung P., Klein H. (2008) Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 [DOI] [PubMed] [Google Scholar]
- 16. Long D. T., Räschle M., Joukov V., Walter J. C. (2011) Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science 333, 84–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Petermann E., Orta M. L., Issaeva N., Schultz N., Helleday T. (2010) Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell 37, 492–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Somyajit K., Subramanya S., Nagaraju G. (2010) RAD51C. A novel cancer susceptibility gene is linked to Fanconi anemia and breast cancer. Carcinogenesis 31, 2031–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nagaraju G., Odate S., Xie A., Scully R. (2006) Differential regulation of short and long tract gene conversion between sister chromatids by Rad51C. Mol. Cell. Biol. 26, 8075–8086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Badie S., Liao C., Thanasoula M., Barber P., Hill M. A., Tarsounas M. (2009) RAD51C facilitates checkpoint signaling by promoting CHK2 phosphorylation. J. Cell Biol. 185, 587–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scully R., Livingston D. M. (2000) In search of the tumor-suppressor functions of BRCA1 and BRCA2. Nature 408, 429–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Venkitaraman A. R. (2002) Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108, 171–182 [DOI] [PubMed] [Google Scholar]
- 23. Scully R., Chen J., Plug A., Xiao Y., Weaver D., Feunteun J., Ashley T., Livingston D. M. (1997) Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88, 265–275 [DOI] [PubMed] [Google Scholar]
- 24. Chen J., Silver D. P., Walpita D., Cantor S. B., Gazdar A. F., Tomlinson G., Couch F. J., Weber B. L., Ashley T., Livingston D. M., Scully R. (1998) Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol. Cell 2, 317–328 [DOI] [PubMed] [Google Scholar]
- 25. Scully R., Chen J., Ochs R. L., Keegan K., Hoekstra M., Feunteun J., Livingston D. M. (1997) Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell 90, 425–435 [DOI] [PubMed] [Google Scholar]
- 26. Lomonosov M., Anand S., Sangrithi M., Davies R., Venkitaraman A. R. (2003) Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev. 17, 3017–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moynahan M. E., Chiu J. W., Koller B. H., Jasin M. (1999) Brca1 controls homology-directed DNA repair. Mol. Cell 4, 511–518 [DOI] [PubMed] [Google Scholar]
- 28. Moynahan M. E., Pierce A. J., Jasin M. (2001) BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell 7, 263–272 [DOI] [PubMed] [Google Scholar]
- 29. Jensen R. B., Carreira A., Kowalczykowski S. C. (2010) Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467, 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Greenberg R. A., Sobhian B., Pathania S., Cantor S. B., Nakatani Y., Livingston D. M. (2006) Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 20, 34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Deans A. J., West S. C. (2011) DNA interstrand cross-link repair and cancer. Nat. Rev. Cancer 11, 467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kee Y., D'Andrea A. D. (2010) Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev. 24, 1680–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Garcia-Higuera I., Kuang Y., Denham J., D'Andrea A. D. (2000) The Fanconi anemia proteins FANCA and FANCG stabilize each other and promote the nuclear accumulation of the Fanconi anemia complex. Blood 96, 3224–3230 [PubMed] [Google Scholar]
- 34. Smogorzewska A., Matsuoka S., Vinciguerra P., McDonald E. R., 3rd, Hurov K. E., Luo J., Ballif B. A., Gygi S. P., Hofmann K., D'Andrea A. D., Elledge S. J. (2007) Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129, 289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ali A. M., Singh T. R., Meetei A. R. (2009) FANCM-FAAP24 and FANCJ. FA proteins that metabolize DNA. Mutat. Res. 668, 20–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Taniguchi T., Garcia-Higuera I., Andreassen P. R., Gregory R. C., Grompe M., D'Andrea A. D. (2002) S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood 100, 2414–2420 [DOI] [PubMed] [Google Scholar]
- 37. Wang X., Andreassen P. R., D'Andrea A. D. (2004) Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol. Cell. Biol. 24, 5850–5862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vaz F., Hanenberg H., Schuster B., Barker K., Wiek C., Erven V., Neveling K., Endt D., Kesterton I., Autore F., Fraternali F., Freund M., Hartmann L., Grimwade D., Roberts R. G., Schaal H., Mohammed S., Rahman N., Schindler D., Mathew C. G. (2010) Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat. Genet. 42, 406–409 [DOI] [PubMed] [Google Scholar]
- 39. Meindl A., Hellebrand H., Wiek C., Erven V., Wappenschmidt B., Niederacher D., Freund M., Lichtner P., Hartmann L., Schaal H., Ramser J., Honisch E., Kubisch C., Wichmann H. E., Kast K., Deissler H., Engel C., Müller-Myhsok B., Neveling K., Kiechle M., Mathew C. G., Schindler D., Schmutzler R. K., Hanenberg H. (2010) Germ line mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat. Genet. 42, 410–414 [DOI] [PubMed] [Google Scholar]
- 40. Puget N., Knowlton M., Scully R. (2005) Molecular analysis of sister chromatid recombination in mammalian cells. DNA Repair 4, 149–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rothfuss A., Grompe M. (2004) Repair kinetics of genomic interstrand DNA cross-links. Evidence for DNA double strand break-dependent activation of the Fanconi anemia/BRCA pathway. Mol. Cell. Biol. 24, 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Olive P. L., Wlodek D., Banáth J. P. (1991) DNA double strand breaks measured in individual cells subjected to gel electrophoresis. Cancer Res. 51, 4671–4676 [PubMed] [Google Scholar]
- 43. Kennedy R. D., D'Andrea A. D. (2005) The Fanconi anemia/BRCA pathway. New faces in the crowd. Genes Dev. 19, 2925–2940 [DOI] [PubMed] [Google Scholar]
- 44. Joenje H., Patel K. J. (2001) The emerging genetic and molecular basis of Fanconi anemia. Nat. Rev. Genet. 2, 446–457 [DOI] [PubMed] [Google Scholar]
- 45. Kuznetsov S., Pellegrini M., Shuda K., Fernandez-Capetillo O., Liu Y., Martin B. K., Burkett S., Southon E., Pati D., Tessarollo L., West S. C., Donovan P. J., Nussenzweig A., Sharan S. K. (2007) RAD51C deficiency in mice results in early prophase I arrest in males and sister chromatid separation at metaphase II in females. J. Cell Biol. 176, 581–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Petermann E., Helleday T. (2010) Pathways of mammalian replication fork restart. Nat. Rev. Mol. Cell Biol. 11, 683–687 [DOI] [PubMed] [Google Scholar]
- 47. Huang J., Huen M. S., Kim H., Leung C. C., Glover J. N., Yu X., Chen J. (2009) RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat. Cell Biol. 11, 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lio Y. C., Mazin A. V., Kowalczykowski S. C., Chen D. J. (2003) Complex formation by the human Rad51B and Rad51C DNA repair proteins and their activities in vitro. J. Biol. Chem. 278, 2469–2478 [DOI] [PubMed] [Google Scholar]
- 49. Rodrigue A., Lafrance M., Gauthier M. C., McDonald D., Hendzel M., West S. C., Jasin M., Masson J. Y. (2006) Interplay between human DNA repair proteins at a unique double strand break in vivo. EMBO J. 25, 222–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bartek J., Lukas C., Lukas J. (2004) Checking on DNA damage in S phase. Nat. Rev. Mol. Cell Biol. 5, 792–804 [DOI] [PubMed] [Google Scholar]
- 51. Taniguchi T., Garcia-Higuera I., Xu B., Andreassen P. R., Gregory R. C., Kim S. T., Lane W. S., Kastan M. B., D'Andrea A. D. (2002) Convergence of the Fanconi anemia and ataxia telangiectasia signaling pathways. Cell 109, 459–472 [DOI] [PubMed] [Google Scholar]
- 52. Takemura H., Rao V. A., Sordet O., Furuta T., Miao Z. H., Meng L., Zhang H., Pommier Y. (2006) Defective Mre11-dependent activation of Chk2 by ataxia telangiectasia mutated in colorectal carcinoma cells in response to replication-dependent DNA double strand breaks. J. Biol. Chem. 281, 30814–30823 [DOI] [PubMed] [Google Scholar]
- 53. Buscemi G., Savio C., Zannini L., Miccichè F., Masnada D., Nakanishi M., Tauchi H., Komatsu K., Mizutani S., Khanna K., Chen P., Concannon P., Chessa L., Delia D. (2001) Chk2 activation dependence on Nbs1 after DNA damage. Mol. Cell. Biol. 21, 5214–5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu Y., Tarsounas M., O'regan P., West S. C. (2007) Role of RAD51C and XRCC3 in genetic recombination and DNA repair. J. Biol. Chem. 282, 1973–1979 [DOI] [PubMed] [Google Scholar]
- 55. Huen M. S., Sy S. M., Chen J. (2010) BRCA1 and its toolbox for the maintenance of genome integrity. Nat. Rev. Mol. Cell Biol. 11, 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gildemeister O. S., Sage J. M., Knight K. L. (2009) Cellular redistribution of Rad51 in response to DNA damage. Novel role for Rad51C. J. Biol. Chem. 284, 31945–31952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. French C. A., Tambini C. E., Thacker J. (2003) Identification of functional domains in the RAD51L2 (RAD51C) protein and its requirement for gene conversion. J. Biol. Chem. 278, 45445–45450 [DOI] [PubMed] [Google Scholar]
- 58. Tarsounas M., Davies D., West S. C. (2003) BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene 22, 1115–1123 [DOI] [PubMed] [Google Scholar]
- 59. Moldovan G. L., D'Andrea A. D. (2009) How the Fanconi anemia pathway guards the genome. Annu. Rev. Genet. 43, 223–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Patel K. J., Yu V. P., Lee H., Corcoran A., Thistlethwaite F. C., Evans M. J., Colledge W. H., Friedman L. S., Ponder B. A., Venkitaraman A. R. (1998) Involvement of Brca2 in DNA repair. Mol. Cell 1, 347–357 [DOI] [PubMed] [Google Scholar]
- 61. Venkitaraman A. R. (2004) Tracing the network connecting BRCA and Fanconi anaemia proteins. Nat. Rev. Cancer 4, 266–276 [DOI] [PubMed] [Google Scholar]
- 62. Xu B., Kim S.t., Kastan M. B. (2001) Involvement of Brca1 in S-phase and G2-phase checkpoints after ionizing irradiation. Mol. Cell. Biol. 21, 3445–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Xu B., Kim S. T., Lim D. S., Kastan M. B. (2002) Two molecularly distinct G2/M checkpoints are induced by ionizing irradiation. Mol. Cell. Biol. 22, 1049–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kraakman-van der Zwet M., Overkamp W. J., van Lange R. E., Essers J., van Duijn-Goedhart A., Wiggers I., Swaminathan S., van Buul P. P., Errami A., Tan R. T., Jaspers N. G., Sharan S. K., Kanaar R., Zdzienicka M. Z. (2002) Brca2 (XRCC11) deficiency results in radioresistant DNA synthesis and a higher frequency of spontaneous deletions. Mol. Cell. Biol. 22, 669–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Xia B., Sheng Q., Nakanishi K., Ohashi A., Wu J., Christ N., Liu X., Jasin M., Couch F. J., Livingston D. M. (2006) Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell 22, 719–729 [DOI] [PubMed] [Google Scholar]
- 66. Nagaraju G., Hartlerode A., Kwok A., Chandramouly G., Scully R. (2009) XRCC2 and XRCC3 regulate the balance between short and long tract gene conversions between sister chromatids. Mol. Cell. Biol. 29, 4283–4294 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.