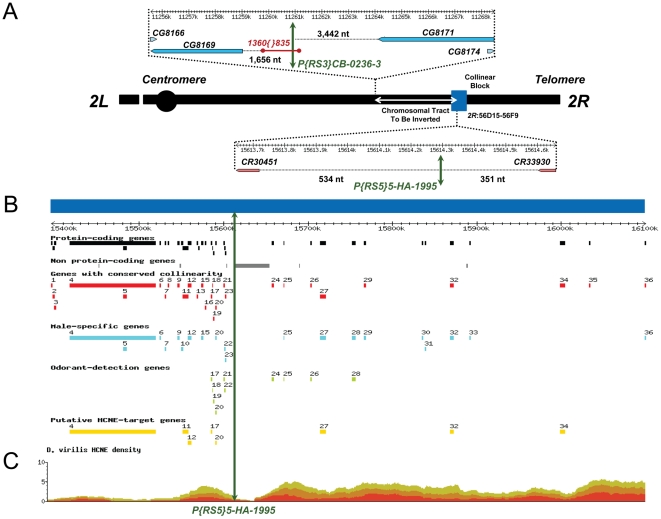
Figure 1. Schematic representation of the inversion engineered to disrupt the ultraconserved region CG15121–CG16894 and its salient features.
(A) Schematic representation showing the surrounding regions and location of the FRT-bearing TEs P{RS5}5-HA-1995 and P{RS3}CB-0236-3 (green double arrowhead lines) used to generate the inversion In(2R)51F11-56E2 (white double arrowhead line), which disrupts the ultraconserved region CG15121–CG16894 (blue box). The element P{RS3}CB-0236-3 is inserted in a naturally occurring copy of the TE 1360{}835 [43]. Distances between the FRT-bearing TEs and the flanking genes are indicated in nucleotides. Notice that flanking transcription units at the immediate vicinity of the inner breakpoint are two non-protein-coding genes: CR30451, which codes for tRNA:E4:56Fc; and CR33930, which codes for snoRNA:185. (B) Annotation of the ultraconserved region CG15121–CG16894 using D. melanogaster as a reference. From top to bottom: 36 protein-coding genes; 122 non-protein-coding genes (8 miRNAs, 13 tRNAs, 1 snoRNA, and 100 5S rRNAs); 29 protein-coding genes whose collinearity is maintained across nine species of the genus Drosophila; 20 protein-coding genes with male-biased gene expression using mRNA levels as a proxy; 10 protein-coding genes related to odor-guided behavior; and 8 putative targets of Highly Conserved Non-coding Elements (HCNEs) based on their expression profile, core promoter predictions, and mutant phenotypes. Details on the protein-coding genes in this region and their annotation are provided in Table S2. (C) Ancora [82] snapshot (http://ancora.genereg.net) of the distribution of HCNEs when genome sequences of D. melanogaster and D. virilis are compared. Green, orange, and red denote 96%, 98%, and 100% nucleotide identity, respectively.