Abstract
Non-structural protein 1 (NS1) of influenza A viruses is a multifunctional protein that antagonizes the host immune response by interfering with several host signaling pathways. Based on putative amino acid sequences, NS1 proteins are categorized into two gene pools, allele A and allele B. Here we identified that allele A NS1 proteins of H6N8 and H4N6 are able to inhibit double-stranded RNA (dsRNA)-induced activating protein-1 (AP-1) promoter in cultured cell lines (human A549 and mink lung cells). Allele B NS1 proteins from corresponding subtypes of influenza A viruses are weak in this inhibition, despite significant levels of expression of each NS1 protein in human A549 cells. Furthermore, the capability to inhibit AP-1 promoter was mapped in the effector domain, since RNA binding domain alone lost its ability to inhibit this promoter activation. Chimeric forms of NS1 protein, composed of either RNA binding domain of allele A or B and effector domain of allele A or B, showed comparable inhibition to that of their wild-type NS1 proteins, or to the effector domain of corresponding NS1 proteins. Both alleles A and B NS1 proteins of H6N8 and H4N6 were expressed to significant levels, and were localized predominantly in the nucleus of human A549 cells. These results underscore the importance of the effector domain in inhibiting AP-1 promoter activation, and the biological function of the effector domain in stabilizing the RNA binding domain. Further, we revealed the versatile nature of NS1 in inhibiting the AP-1 transcription factor, in a manner dependent on allele type. Comprehensive studies, focusing on the molecular mechanisms behind this differential inhibition, may facilitate exploration of the zoonotic and pathogenic potential of influenza A viruses.
Introduction
Mammalian cells initiate an innate immune response, primarily in the form of alpha/beta interferons (IFN-α/β) as a first line of protection against invading pathogens. The synthesis and secretion of IFN-α/β requires activated and constitutively expressed transcription factors. Although IFN-α/β mRNA transcription is independent of de novo protein synthesis, it occurs via virus-induced activation of transcription factors. In uninfected cells, three transcription factors, nuclear factor-κB (NF-κB), interferon regulatory factor-3/7 (IRF-3/7), and activating protein-1 (AP-1), reside in the cytoplasm, which upon activation translocate to the nucleus, where they initiate IFN-α/β transcription by recruiting the transcriptional coactivator CREB-binding protein (CBP) (20). The AP-1 transcription factor is composed of Jun (c-Jun, JunB, and JunD), Fos (c-Fos, FosB, Fra-1, and Fra-2), or the activating transcription factor (ATF-2 and ATF-3) proteins, and these proteins bind to a common specific positive regulatory domain IV (PRD IV). IRF-3 binds to PRD III and I, whereas NF-κB binds to PRD II within the IFN-β promoter region (7). The integrated assembly of enhanceosome on these PRDs is essential for the maximal activation of IFN-β promoter (12). Intracellular double-stranded RNA (dsRNA) such as viral by-product or poly I:C, initiates a complex set of pathways that culminate in the activation of AP-1 via Jun N-terminal kinase (JNK), along with other transcription factors (5,9). The non-structural protein 1 (NS1) of influenza A viruses is a multifunctional protein that inhibits AP-1 transcription factor as a strategy to subvert the host immune response (10).
The NS1 protein of influenza A viruses is highly conserved and structurally divided into two domains, joined by a flexible linker (3). The first 73 residues in the N-terminal constitute a double-stranded RNA binding domain (RBD), whereas residues from 86–230 aa make up an effector domain (ED). RBD binds non-specifically to dsRNA, and is mainly responsible for the inhibition of the interferon-induced 2′-5′ oligo A synthetase/RNase L pathway (13). The ED interacts with numerous host cellular proteins such as protein kinase R (PKR) (14), human tripartite motif 25 (TRIM25) (1,15), the p85β subunit of phosphatidylinositol-3-kinase (PI3K) (2), and the 30-kDa subunit of the cleavage and polyadenylation specificity factor (CPSF30) (18,19). Apart from these functions, ED also stabilizes the RBD, which is required for proper functionality of NS1 protein (24). All these functions of NS1 protein consequently result in virulence of influenza infection by antagonizing IFN production and apoptosis.
NS1 is a relatively conserved protein, but there have been two clear divisions based on amino acid sequences, known as alleles A and B (22). It has been noted that allele A comprises strains of both avian and mammalian (human, equine, and swine) origin, whereas allele B includes strains of avian origin with only two exceptions (A/equine/Jilin/1/1989/H3N8 and A/Swine/Saskatchewan/18789/2002/H1N1). In spite of being mammalian origin viruses, these two strains belong to allele B (17).
The role of the NS segment in viral replication has not been investigated conclusively, although it has been demonstrated that A/FPV/Rostock/34 (HPAIV H7N1 with allele A), if carrying allele B NS segment, undergoes attenuation in squirrel monkeys (22). On the other hand, it has been recently observed that A/FPV/Rostock/34, if it has received allele B NS segment from A/Goose/Guangdong/1/96 (H5N1), becomes more virulent than the wild-type H7N1, and can cause infectivity in mice (11,23). The proficient replication of recombinant viruses in the presence of allele B suggests that the NS segment helps the virus cross host barriers, and is fundamental for cell tropism, host range, and pathogenicity determination. Apart from that, our group recently showed that allele B NS1 proteins of different avian influenza A viruses antagonize IFN-β significantly less than allele A NS1 proteins of corresponding subtypes (17,30). NF-κB is a general prerequisite for influenza A virus replication. In this regard, we have shown that allele A NS1 is more potent in dsRNA-induced NF-κB promoter inhibition compared to allele B NS1 proteins of corresponding influenza isolates (16,17). In this report, we focused our investigations on the structural and functional abilities of allele A and B NS1 proteins of avian influenza A viruses in inhibiting dsRNA-induced AP-1 promoter activation in cultured cell lines.
Materials and Methods
Four avian influenza A viruses, two of each carrying allele A and B, were used in the study. The nucleotide sequence of allele A of H6N8 (allele A, A/mallard/Sw/412/05, EU518721), allele B of H6N8 (allele B, A/mallard/Sw/418/05, EU518722), allele A of H4N6 (allele A, A/mallard/Sw/818/05, EU518757), and allele B of H4N6 (allele B, A/mallard/Sw/795/05, EU518749) were submitted to GenBank. First, NS1 genes from all four isolates were cloned into mammalian expression vector pcDNA3.1+ (Invitrogen, Carlsbad, CA). These plasmids express full-length NS1 protein and were named H6N8-A, H6N8-B, H4N6-A, and H4N6-B. To investigate the role of each domain of NS1 protein, RNA-binding domain (H6N8-A-RNA and H6N8-B-RNA) from amino acids (aa) 1–73, and effector domain (H6N8-A-ED and H6N8-B-ED) from aa 74–230, were also cloned as previously described (17). Likewise, chimeric NS1 proteins were constructed (H6N8 chiNS1 A/B and H6N8 chiNS1 B/A), each carrying either RNA binding domain of allele A (1–73 aa), and effector domain of allele B (74–230 aa), or vice versa. The orientation of clones in these constructs was confirmed using DNA sequencing, while TNT (Promega, Fitchburg, WI) in combination with 35S radiolabeling was applied to check expression of all clones.
Two mammalian cell lines, mink lung cells (MiLu) and alveolar epithelial cells from human adenocarcinoma (A549), were maintained in Eagle's minimum essential medium and Dulbecco's modified Eagle's medium, respectively, as previously described (17). To test the AP-1 promoter activation, A549 and MiLu were seeded in 24-well plates with approximately 2.5×104 cells per well. The next day, the 24-well plates were transfected with 500 ng of pAP-1-Luc reporter plasmid (pAP-1-Luc, containing AP-1 promoter tagged with a luciferase gene), and either 500 ng of NS1 expression constructs or empty pcDNA3.1+ plasmid, using FuGENE6 (Roche, Madison, WI). After 24 h, the cells were stimulated with 10 μg/mL of poly I:C (dsRNA; Invivogen) using Lipofectamine 2000 transfection reagent (Invitrogen). At 24 h post-stimulation, the luciferase activity was determined using the ONE-Glo™ Luciferase assay system (Promega) and a Wallac Victor™ 1420 multilabel counter (Wallac Sverige AB, Vasby, Sweden), as directed by the manufacturer. pTA-Luc plasmid was also transfected to eliminate the background of reporter gene activity, and normalized by subtracting from all the samples. The empty pcDNA3.1+ plasmid served as a negative control and was used to normalize the DNA concentration in the reporter control. All transfections were carried out in duplicate, and all experiments were repeated at least three times. The data were analyzed using Student's t-test, and comparisons showing p values <0.05 were designated as significant.
The expression of NS1 proteins in A549 cells was investigated using a highly sensitive assay known as in situ proximity ligation assay (in situ PLA), using a Duolink in situ PLA kit (Olink Biosciences, Uppsala, Sweden), as we previously reported (17). Antibodies to NS1, raised against the C-terminus of NS1 protein, were purchased from ProScie (Poway, CA).
Results
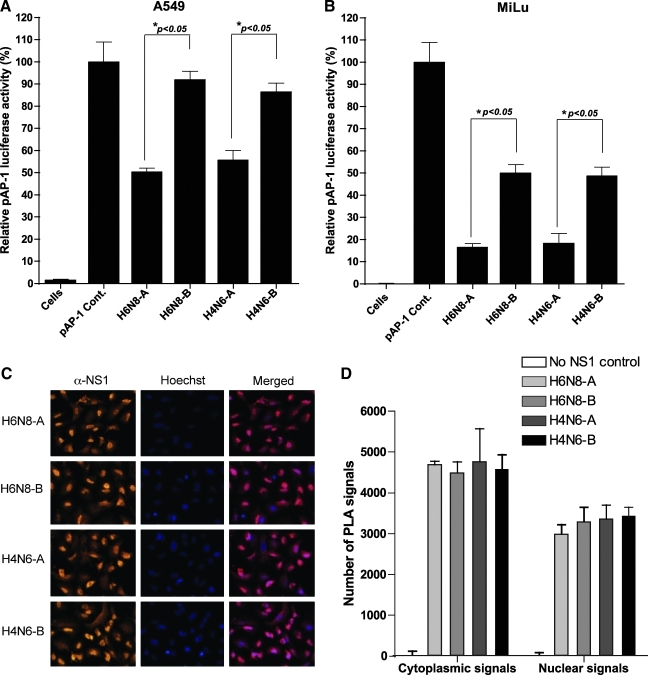
To investigate the function of the NS1 protein of avian influenza A viruses against the induction of AP-1 promoter, we constructed expression vectors that encode for full length NS1 protein derived from allele A and B subtypes of H6N8 and H4N6. Initially we tested whether these NS1 proteins are capable of inhibiting dsRNA-induced AP-1 promoter activation. The expression vectors for allele A and B NS1 proteins of both subtypes of influenza A viruses were co-transfected with pAP-1-Luc reporter plasmids into A549 cells and subjected to dsRNA stimulation before lysis for determination of luciferase activity. The results showed that the cells transfected with AP-1 reported plasmid and empty vector (pAP-1 Cont.) expressed the highest luciferase activity and was set to 100% for comparison purposes after normalization. However, the activation of AP-1 promoter was significantly inhibited by the expression of allele A NS1 proteins from both subtypes (∼52%; p<0.05; Fig. 1A). Notably, the allele B NS1 proteins of corresponding subtypes were unable to inhibit this exogenously administered dsRNA-induced AP-1 promoter activity, although a low-level, statistically non-significant reduction was obvious (H6N8-B=8% and H4N6-B=14%). At this point, the results from the reporter assay demonstrated that allele A NS1 protein inhibits AP-1 promoter activation by interfering in the signal transduction pathway that leads to activation of the AP-1 promoter, whereas allele B NS1 proteins from the corresponding influenza subtypes lack this ability, and hence did not interfere in this signaling pathway.
FIG. 1.
Differential inhibition of AP-1 promoter by allele A and B NS1 proteins of avian influenza A viruses in A549 and MiLu cells, in spite of significant levels of expression in A549 cells. (A) Five hundred nanograms of each NS1 expression plasmid (H6N8-A, H6N8-B, H4N6-A, or H4N6-B) or empty plasmid were co-transfected with 500 ng of pAP-1-Luc reported plasmid in A549 cells. The cells were stimulated by Lipfectamine 2000 transfected dsRNA (10 μg/mL), 24 h post-transfection. After another 24 h, the cells were lysed and luciferase activity was determined. The mock-treated control (pAP-1 Cont.) was set to 100%, and the level of inhibition was reported in percentages. (B) The same experimental setup was repeated for MiLu cells. Error bars indicate standard deviations. The data presented here correspond to three experiments with transfections performed in duplicate (*p<0.05 by Student's t-test). (C) A549 cells were transfected with expression vector encoding NS1 gene for H6N8-A, H6N8-B, H4N6-A, or H4N6-B. The cells were fixed at 18 h post-transfection and were run for in situ PLA. (D) The quantification of cytoplasmic and nuclear signals as measured by BlobFinder software. The data represent means±SD of three microscopic fields. Color images available online at www.liebertonline.com/vim
It was of concern whether the expression of NS1 protein actually affects AP-1 promoter activation in a cell-specific manner. For this purpose, MiLu cells were transfected with NS1 expression plasmids, along with pAP-1-Luc reporter plasmid and stimulated with the transfection of dsRNA. The results showed that both allele A and B NS1 proteins of avian influenza A viruses were strong inhibitors of AP-1 promoter activation (Fig. 1B). However, consistent with the results in A549 cells, allele A NS1 proteins of H6N8 and H4N6 were significantly potent in inhibiting the AP-1 promoter compared with allele B NS1 proteins of corresponding influenza A viruses. Allele B NS1 proteins of both influenza subtypes inhibited the AP-1 promoter at around 50%, whereas allele A NS1 proteins of corresponding isolates showed an inhibition of 86% in MiLu cells (Fig. 1B).
At this point, it was plausible to conclude that allele A and B NS1 proteins of avian influenza A viruses differ in their abilities to block AP-1 pathways, and that allele A is more potent than allele B in this regard. However, the differences in the level of expression of allele B could be a possible cause for this weaker inhibition. To rule out this possibility, we employed a highly sensitive technique (in situ PLA), to check not only the expression of NS1 proteins, but also their nuclear localization pattern in A549 cells (27). NS1 expression constructs from both alleles A and B, belonging to both subtypes, were transfected into A549 cells seeded in 8-well chambers. The cells were fixed at 12 h post-transfection and processed for in situ PLA. As is shown in Fig. 1C, the NS1 proteins from both alleles of H6N8 and H4N6 were expressed efficiently and at comparable levels. Apparently, there was no detectable difference between the two alleles, which was also confirmed by quantitative measurement of signals using BlobFinder software (Fig. 1D). Moreover, at this time point post-transfection, NS1 proteins from both alleles were predominantly localized in the nucleus. Thus, these results demonstrated that the weaker inhibition of the AP-1 promoter under the presence of expressed allele B NS1 protein is due to a functional inability to interfere in the AP-1 pathway, and is not due to differential expression levels in A549 cells.
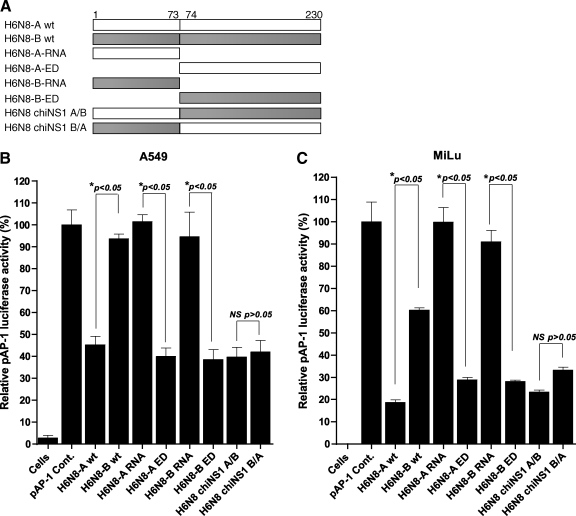
It has been demonstrated that the RNA binding domain is primarily involved in inhibition of interferon production, and that the effecter domain interacts with several cellular proteins. The role of each of these domains has not been investigated with regard to inhibition of the AP-1 pathway. Secondly, the genetic basis behind differential regulation of the AP-1 promoter by allele A and B NS1 proteins requires mapping. For these reasons, the RNA binding domains and effector domains of allele A and B NS1 protein of H6N8 were separately cloned in a mammalian expression vector (Fig. 2A), and tested against AP-1 promoter inhibition in A549 and MiLu cells. In the first set of experiments, constructs for RNA binding and effector domains were transfected into A549 cells along with pAP-1-Luc, and luciferase activity was determined after stimulation with dsRNA. The results, as shown in Fig. 2B, demonstrate that RNA binding domains from both alleles (A and B) were functionally incapable of supporting AP-1 promoter inhibition, and were statistically non-significant compared to a positive control transfected with reporter plasmid, and with DNA concentration normalized with empty vector. This was expected since the RNA binding domain requires dimerization for its functionality. On the other hand, effector domains from both alleles A and B were as strongly inhibitor (60%) of AP-1 promoter activation as that of their wild-type (wt) (55%). The effector domains were significantly better in inhibiting the promoter compared to RNA binding domains (p<0.05). Again, a cross-species comparison in MiLu cells revealed a level corresponding to that of A549 cells. However, the effector domain caused comparatively stronger inhibition of AP-1 promoter (73%) in MiLu cells compared to the inhibition of corresponding domains in A549 cells (Fig. 2C). These structural mappings of NS1 protein against AP-1 promoter inhibition showed that the effector domain is functionally independent of the RNA binding domain, and has the ability to inhibit AP-1 promoter to the same extent as its wild-type, whereas the RNA binding domain alone is unable to support this inhibition.
FIG. 2.
Construction of individual domain and chimeric NS1 gene-containing expression plasmids and investigation of their contribution to the inhibition of AP-1 promoter. (A) A layout for the construction of RNA binding domain, effector domain, and chimeric NS1, for both allele A and B of H6N8. (B) A549 cells were co-transfected with individual domain and chimeric NS1-expressing constructs, along with pAP-1-Luc reporter plasmid, with an equal amount (500 ng). Cells were lysed for luciferase activity determination after stimulation with dsRNA. (C) The same experimental setup was repeated for MiLu cells. The pAP-1 Cont. was normalized with empty vector and was set to 100%. Error bars show standard deviations of the three experiments with transfections performed in duplicate (*p<0.05 by Student's t-test; NS, not significant).
To further confirm these tested functions of RNA binding and effector domains, two chimeric constructs were prepared (as outlined in Fig. 2A). A549 and MiLu cells were transfected with chimeric NS1 expression plasmid and pAP-1-Luc reporter plasmid, and cells were dsRNA-stimulated and then lysed for determination of luciferase activity. As expected, a complete NS1 protein, regardless of the nature of the RNA-binding domain, strongly inhibited AP-1 promoter at comparable levels to that of effector domain alone (MiLu), or effector domain alone and its wild-type (A549; Fig. 2B and C). This reversal of function to inhibit dsRNA-induced AP-1 promoter activation demonstrated that both RNA binding and effector domains are functionally interactive in a co-operative manner, which results in their ability to inhibit the AP-1 promoter, and that RNA-binding domain alone is non-functional in this regard.
Discussion
Activation of the AP-1 pathway is critical to the innate response to a large number of pathogens by initiating production of several antiviral and proinflammatory cytokines. It has been demonstrated that influenza virus infection provokes AP-1 transcription factor-dependent antiviral cytokines such as IFN-β, and that this occurs via JNK phosphorylation (9). Production of IFN-β is an initial event that is responsible for triggering the production of over 300 IFN-stimulated genes, which then collectively establish an antiviral state (20). There are abundant examples of the evasive strategies that different viruses have evolved to antagonize these systems. NS1 protein of avian influenza A viruses is one of the well-characterized proteins implicated in inhibiting IRFs, NF-κB, and the AP-1 transcription family (10,21,25). Due to recent concerns about the differential regulation of IFN-β, for which allele B was found to be weaker in inhibiting IFN-β compared to allele A (17,30), and due to indications that AP-1 is an important regulator of IFN-β (7), we primarily focused on the inhibition of this AP-1 pathway by allele A and B NS1 proteins of influenza A viruses. Furthermore, emphasis was given to structural mapping of the NS1 protein. These would collectively determine if NS1 acts as an important contributor to the clinical outcome and pathogenic potential of these influenza viruses.
Studies have demonstrated that some influenza A viruses with NS1 protein of allele B type have a tendency to replicate efficiently compared to their naïve or recombinant forms (carrying allele A NS1 or delNS1) (11,23). Although the exact mechanism behind this tendency is ill-defined, studies have shown that allele B NS segment-carrying influenza viruses show increased accumulation of viral mRNA compared to allele A, under the same genetic background (27). This suggests that allele B facilitates the transcription and replication of influenza A virus infection by providing a surplus of viral mRNA. The activated NF-κB protects the cells from apoptosis, and influenza virus titer is found to be inversely correlated to the level of apoptosis (27). Likewise, an activated NF-κB is a general prerequisite for efficient viral replication, and NS1 protein inhibits hyper-activation of NF-κB to prevent the NF-κB-mediated antiviral response (25); this function of NS1 was also recently revealed to be allele dependent (16). Furthermore, high putative amino acid differences (∼31%) between allele A and B NS1 protein draw our attention to a possible correlation to the pathogenicity of influenza viruses, because the NS gene segment has been declared an essential factor for cell tropism, host range, and pathogenicity determination (11).
As is the case for viral infection, exogenously administered dsRNA analog (poly I:C) proved to be a strong inducer of AP-1 promoter when transfected into the cells. It has previously been shown that dsRNA induces JNK, which allows us to hypothesize that dsRNA activates AP-1 promoter via JNK phosphorylation (5). However, the results indicate that none of the NS1 protein from any isolate inhibits dsRNA-induced c-Jun phosphorylation at site 239 (data not shown). Given that AP-1 transcription consists of several subunits (7), either inhibition of c-Jun phosphorylation at site 239 is not NS1 dependent, or the inhibition was not sufficient to result in a detectable difference in the immunofluorescence assay. A certain level of AP-1 promoter activity in both allele A and B NS1 proteins further justifies this hypothesis.
In this report, we were able to confirm that NS1 proteins of avian influenza A viruses (H6N8 and H4N6) have an ability to inhibit dsRNA-induced AP-1 transcription factor, and that this property of NS1 is allele-dependent, in the presence of significant amounts of NS1 protein expression in both allele A and B NS1 proteins. The prevalence of the H6 and H4 subtypes of influenza viruses is relatively high in wild bird populations, and they have been isolated from seals, poultry, and swine (4,6,28). Although these viruses are considered to be of low pathogenicity, they are of growing importance for both avian and mammalian species. On the other hand, it has been demonstrated that most of the highly pathogenic influenza A viruses belong to the allele A NS1 group, and share approximately 68% identity with the allele B NS1 group of corresponding subtypes (29). Furthermore, the NS1 protein of H5N1 and H7N1 subtypes of highly pathogenic influenza A virus (HPAIV) has been found to be essential for virulence, host range, and cell tropism (8,26). The effects of allele A and allele B NS1 proteins on the pathogenesis of HPAIV demand further investigation.
Structural mapping further demonstrated that it is the C-terminal effector domain of NS1 protein that is actually responsible for this inhibition, regardless of the nature of the RNA-binding domain. Inability of the RNA-binding domain to inhibit AP-1 promoter may best be explained by the requirement of dimerization of domain (24). Since the effector domain is not only responsible for interaction with cellular proteins, but also facilitates the dimerization of the RNA-binding domain, it is plausible to assume that the RNA-binding domain without effector domain lost its ability to be dimerized, and hence affected its three-dimensional structure, and subsequently lost its functionality. Upon comparison of allele A and B NS1 protein sequences of H6N8 and H4N6 (17), notable differences were found at position 166 (L in allele A and M in allele B), which is characterized by an interaction with P13K (29), and at position 221 (K in allele A and Y in allele B), which is an essential amino acid for regulating nuclear localization (8). Moreover, the presence of a stretch of eight amino acids (21RFADQELG28 and 21LLSMRDMC28) in alleles A and B, respectively, in the middle of the RNA-binding domain warrants further investigation of their biological significance.
In summary, our results demonstrate that NS1 proteins of avian influenza A viruses (H6N8 and H4N6) differentially inhibit the dsRNA-induced activation of AP-1 promoter, an important transcription factor for triggering antiviral cytokine production, which is an important factor in the host's defense mechanisms. Thus we examined the versatile nature of NS1 in inhibiting the AP-1 transcription factor. An understanding on a molecular level of this pathway may lead to a better understanding of the zoonotic and pathogenic potential of influenza A viruses, and may lead to the development of alternative strategies to mitigate influenza virus infections in various host species.
Acknowledgments
This work was supported by a grant from the Swedish Research Council for the Environment, Agricultural Sciences and Spatial Planning (Formas grant 221-2007-935). S.B. is the recipient of the Award of Excellence (Excellensbidrag) of the Swedish University of Agricultural Sciences (SLU). and M.M. is recipient of a fellowship from the Higher Education Commission of Pakistan. The authors would also like to thank Jenna Anderson for English proofreading of this article.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Gack MU. Albrecht RA. Urano T, et al. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hale BG. Jackson D. Chen YH. Lamb RA. Randall RE. Influenza A virus NS1 protein binds p85beta and activates phosphatidylinositol-3-kinase signaling. Proc Natl Acad Sci USA. 2006;103:14194–14199. doi: 10.1073/pnas.0606109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hale BG. Randall RE. Ortin J. Jackson D. The multifunctional NS1 protein of influenza A viruses. J Gen Virol. 2008;89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- 4.Hinshaw VS. Bean WJ. Webster RG, et al. Are seals frequently infected with avian influenza viruses? J Virol. 1984;51:863–865. doi: 10.1128/jvi.51.3.863-865.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iordanov MS. Paranjape JM. Zhou A, et al. Activation of p38 mitogen-activated protein kinase and c-Jun NH(2)-terminal kinase by double-stranded RNA and encephalomyocarditis virus: involvement of RNase L, protein kinase R, and alternative pathways. Mol Cell Biol. 2000;20:617–627. doi: 10.1128/mcb.20.2.617-627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karasin AI. Brown IH. Carman S. Olsen CW. Isolation and characterization of H4N6 avian influenza viruses from pigs with pneumonia in Canada. J Virol. 2000;74:9322–9327. doi: 10.1128/jvi.74.19.9322-9327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karin M. Liu Z. Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 8.Li Z. Jiang Y. Jiao P, et al. The NS1 gene contributes to the virulence of H5N1 avian influenza viruses. J Virol. 2006;80:11115–111123. doi: 10.1128/JVI.00993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ludwig S. Ehrhardt C. Neumeier ER. Kracht M. Rapp UR. Pleschka S. Influenza virus-induced AP-1-dependent gene expression requires activation of the JNK signaling pathway. J Biol Chem. 2001;276:10990–10998. [PubMed] [Google Scholar]
- 10.Ludwig S. Wang X. Ehrhardt C, et al. The influenza A virus NS1 protein inhibits activation of Jun N-terminal kinase and AP-1 transcription factors. J Virol. 2002;76:11166–11171. doi: 10.1128/JVI.76.21.11166-11171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma W. Brenner D. Wang Z, et al. The NS segment of an H5N1 highly pathogenic avian influenza virus (HPAIV) is sufficient to alter replication efficiency, cell tropism, and host range of an H7N1 HPAIV. J Virol. 2010;84:2122–2133. doi: 10.1128/JVI.01668-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maniatis T. Falvo JV. Kim TH. Kim TK. Lin CH. Parekh BS. Wathelet MG. Structure and function of the interferon-beta enhanceosome. Cold Spring Harb Symp Quant Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 13.Min JY. Krug RM. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2′-5′ oligo (A) synthetase/RNase L pathway. Proc Natl Acad Sci USA. 2006;103:7100–7105. doi: 10.1073/pnas.0602184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Min JY. Li S. Sen GC. Krug RM. A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology. 2007;363:236–243. doi: 10.1016/j.virol.2007.01.038. [DOI] [PubMed] [Google Scholar]
- 15.Munir M. TRIM proteins: another class of viral victims. Sci Signal. 2010;3:jc2. doi: 10.1126/scisignal.3118jc2. [DOI] [PubMed] [Google Scholar]
- 16.Munir M. Zohari S. Berg M. Non-structural protein 1 of avian influenza A viruses differentially inhibit NF-kappaB promoter activation. Virol J. 2011;8:383. doi: 10.1186/1743-422X-8-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munir M. Zohari S. Metreveli G. Baule C. Belak S. Berg M. Allele A and B non-structural protein 1 of avian influenza A viruses differentially inhibit IFN-beta production in human and mink lung cells. J Gen Virol. 2011;92:2111–2121. doi: 10.1099/vir.0.031716-0. [DOI] [PubMed] [Google Scholar]
- 18.Nemeroff ME. Barabino SM. Li Y. Keller W. Krug RM. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol Cell. 1998;1:991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 19.Noah DL. Twu KY. Krug RM. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAS. Virology. 2003;307:386–395. doi: 10.1016/s0042-6822(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 20.Randall RE. Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 21.Talon J. Horvath CM. Polley R. Basler CF. Muster T. Palese P. Garcia-Sastre A. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J Virol. 2000;74:7989–7996. doi: 10.1128/jvi.74.17.7989-7996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Treanor JJ. Snyder MH. London WT. Murphy BR. The B allele of the NS gene of avian influenza viruses, but not the A allele, attenuates a human influenza A virus for squirrel monkeys. Virology. 1989;171:1–9. doi: 10.1016/0042-6822(89)90504-7. [DOI] [PubMed] [Google Scholar]
- 23.van Wielink R. Harmsen MM. Martens DE. Peeters BP. Wijffels RH. Moormann RJ. MDCK cell line with inducible allele B NS1 expression propagates delNS1 influenza virus to high titres. Vaccine. 2011;40:6976–6985. doi: 10.1016/j.vaccine.2011.07.037. [DOI] [PubMed] [Google Scholar]
- 24.Wang X. Basler CF. Williams BR. Silverman RH. Palese P. Garcia-Sastre A. Functional replacement of the carboxy-terminal two-thirds of the influenza A virus NS1 protein with short heterologous dimerization domains. J Virol. 2002;76:12951–12962. doi: 10.1128/JVI.76.24.12951-12962.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X. Li M. Zheng H. Muster T. Palese P. Beg AA. Garcia-Sastre A. Influenza A virus NS1 protein prevents activation of NF-kappaB and induction of alpha/beta interferon. J Virology. 2000;74:11566–11573. doi: 10.1128/jvi.74.24.11566-11573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z. Robb NC. Lenz E. Wolff T. Fodor E. Pleschka S. NS reassortment of an H7-type highly pathogenic avian influenza virus affects its propagation by altering the regulation of viral RNA production and antiviral host response. J Virol. 2010;84:11323–11335. doi: 10.1128/JVI.01034-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weibrecht I. Leuchowius KJ. Clausson CM. Conze T. Jarvius M. Howell WM. Kamali-Moghaddam M. Soderberg O. Proximity ligation assays: a recent addition to the proteomics toolbox. Expert Rev Proteomics. 2010;7:401–409. doi: 10.1586/epr.10.10. [DOI] [PubMed] [Google Scholar]
- 28.Wisedchanwet T. Wongphatcharachai M. Boonyapisitsopa S. Bunpapong N. Kitikoon P. Amonsin A. Genetic characterization of avian influenza subtype H4N6 and H4N9 from live bird market, Thailand. Virol J. 2011;8:131. doi: 10.1186/1743-422X-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zohari S. Gyarmati P. Thoren P. Czifra G. Brojer C. Belak S. Berg M. Genetic characterization of the NS gene indicates co-circulation of two sub-lineages of highly pathogenic avian influenza virus of H5N1 subtype in Northern Europe in 2006. Virus Genes. 2008;36:117–125. doi: 10.1007/s11262-007-0188-7. [DOI] [PubMed] [Google Scholar]
- 30.Zohari S. Munir M. Metreveli G. Belak S. Berg M. Differences in the ability to suppress interferon beta production between allele A and allele B NS1 proteins from H10 influenza A viruses. Virol J. 2010;7:376. doi: 10.1186/1743-422X-7-376. [DOI] [PMC free article] [PubMed] [Google Scholar]