Figure 2.
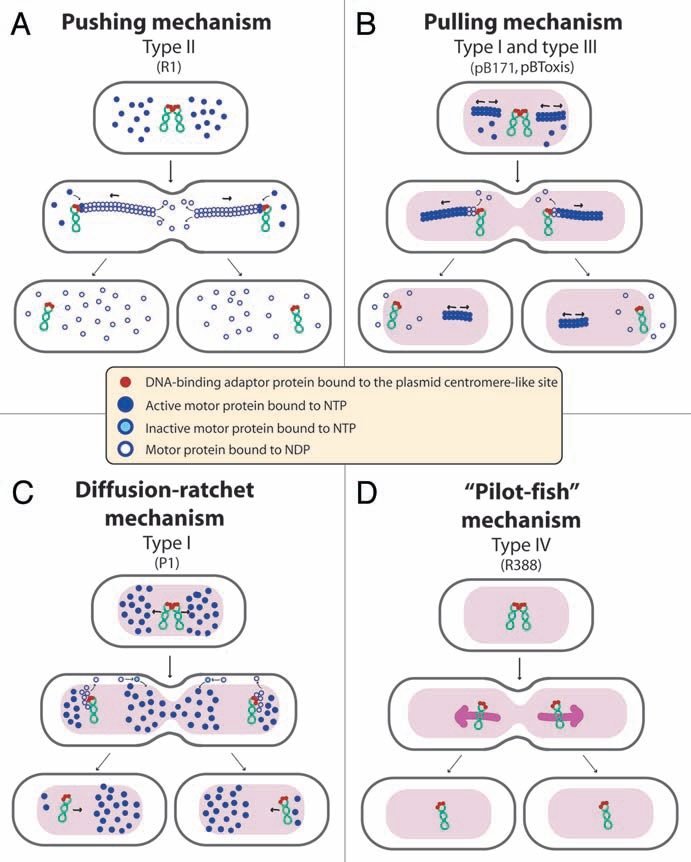
Proposed mechanisms of plasmid segregation. For the sake of clarity, only two newly replicated plasmid copies are represented. When necessary, light red and white regions indicate nucleoid and cytosol spaces, respectively. (A) Pushing mechanism, exemplified by R1 type II par system. Partitioning complexes are formed through specific binding of ParR proteins (red circles) to the centromere-like site parC of newly replicated plasmid molecules, and serve as a nucleation point for ParM-mediated filament formation. Continuous insertion of ParM-ATP motor proteins (blue circles) on the filament ends pushes plasmid molecules apart. Conversion of ParM-ATP to ParM-ADP (open blue circles) leads to destabilization of the filament, thus allowing the entry of another ParM-ATP. At cell division, plasmid molecules localize near opposite cell poles, thus ending in daughter cells.15 (B) Pulling mechanism, as proposed for type I (pB171) and type III (pBtoxis) par systems.16,17 NTP-bound motor proteins (blue circles) bind cooperatively to nucleoid DNA, forming a nucleating core from which filaments form. Subsequently, a growing filament contacts a partition complex formed by the adaptor protein (red circles) bound to the plasmid centromere-like site. Stimulation of NTPase activity of the motor protein by the adaptor protein at the end of the filament leads to conversion to its NDP form (open blue circles) and its release, leaving a new filament end accessible for interactions with the partition complex. The plasmid is thus pulled in the opposite direction to the growth of the filament, and moves around its position, between two other plasmids or between a plasmid and the nucleoid end. (C) Diffusion-ratchet mechanism, as proposed for plasmid P1 type I par system.18 ParB (red circles) loads onto the plasmid at the centromere-like site parS, forming the partition complex. After plasmid replication, partition complexes develop repulsive interactions. ParB stimulates ParA (dark blue circles) ATPase activity, and ParA-ADP molecules (open blue circles) are then excluded from the nucleoid. The motive force for plasmid movement is directed toward regions of high ParA concentration. Movement of the partition complex is thus constrained to one direction because of the low ParA concentration behind it, and at the nucleoid end, it changes direction. ParA-ADP molecules diffuse randomly, exchange ADP for ATP (light blue circles), and then rebind the nucleoid. (D) “Pilot-fish” mechanism, as proposed for plasmid R388 segregation, representing the prototype of a potentially new class of segregation system type IV.21 In contrast to typical par systems, the StbB putative ATPase is not involved in R388 segregation, which does not need a motor protein. We propose that the partition complex, formed by StbA binding to the centromere-like site stbDRs, is used to pair plasmid molecules to the host nucleoid (or other structure associated to the nucleoid). Plasmid segregation is ensured by the host chromosome segregation system.
