Table 1. Sites under PDS in the A. thaliana MADS-box gene family: “Site-specific analyses”.
| Site-specific analyses (type 1) | n | dN/dS (ω) under M0 |
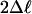 M3 vs. M0 (df LRT 3) M3 vs. M0 (df LRT 3) |
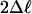 M2 vs. M1 (df 2) M2 vs. M1 (df 2) |
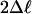 M8 vs. M7 (df 2) M8 vs. M7 (df 2) |
Parameter estimates (β and ω) under M8 β (p,q) | Positively selected sites under M3 | Positively selected sites under M2 | Positively selected sites under M8 |
|---|---|---|---|---|---|---|---|---|---|
| Node AG | 7 | 0.22 | 28.76*** | 10.10* | 10.57* | P1 = 0.98, ω = 8.44 β (0.87, 3,82) | 123 | No rate classes with dN/dS > 1 | 123 |
| Node AH | 7 | 0.26 | 59.09*** | 11.75** | 7.36* | P1 = 0.81, ω = 1.96 β (0.87, 3.48) | 73 7461 72 82 107 63 | 73 747261 82 63 107 | 73 72 7461 82 10763 48 58 81 110 |
| Node AL | 9 | 0.25 | 90.35*** | 18.39*** | 4.55 | P1 = 0.93, ω = 1.24 β (0.80, 2.28) | 61 72 73 82 107 110 48 74 8658 64 6663 65 109 | 72 73 827461 107 48 110 | 7273 8248 61 74 107 110 58 |
| Node AP | 12 | 0.25 | 130.47*** | 21.26*** | 7.59* | P1 = 0.75, ω = 1.18 β (0.56, 3.46) | 58 | 58 48 57 78 8972 12373 81 | 48 57 58 72 73 78 89 123 59 65 8147 6066 92 74 100 |
| Node AQ | 14 | 0.22 | 116.71*** | 14.97*** | 7.73* | P1 = 0.97, ω = 4.67 β (0.47, 1.52) | 5889 | 58 | 58 |
| Node AR | 20 | 0.15 | 102.65*** | 30.18*** | 8.22* | P1 = 0.96, ω = 1.12 β (1.64, 8.94) | No rate classes with dN/dS > 1. | 73 82 72 | 82 73 |
Each comparison has n sequences, dN/dS is average ratio over sites under a codon model with one ω. Proportion of the component of positively selected sites (P1) and parameters p and q of the beta distribution β(p,q) are given under M8. *, P < 0.05; **, P < 0.005; ***, P < 0.001; bold underlined, PP ≥ 0.99 of being under positive selection; bold, 0.99 > PP ≥ 0.95; italics, 0.95 > PP ≥ 0.90; underlined, 0.90 > PP ≥ 0.70; normal, 0.70 > PP ≥ 0.50.
