Figure 2.
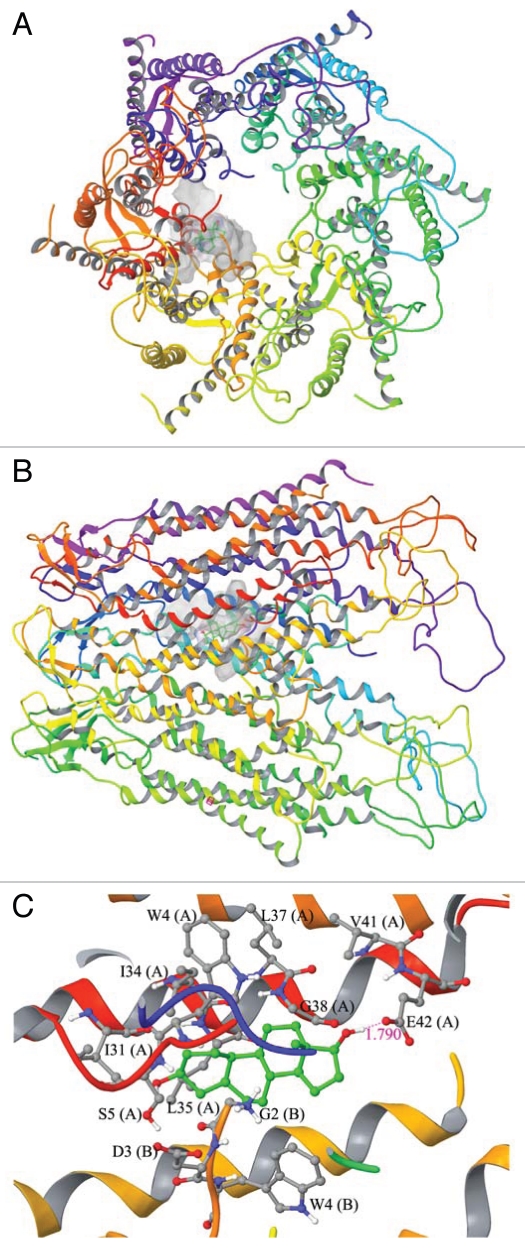
A molecular modeling study to assess the docked complex of estradiol-17β with the Cx43-based gap junction communication channel. (A) Topographic view of the Cx43-gap junction protein complex with estradiol-17β. (B) Side view of the Cx43-gap junction protein complex with estradiol-17β. (C) Ball and stick model for the interaction residues and secondary structure representation for protein between estradiol-17β and Cx43. The SOSUI WWW server65 was used to predict transmembrane helices for connexin 43 (Cx43)-based gap junction (GJ) communicating channel. The characteristic feature of the Cx43-GJ channel is the spanning of transmembrane helices in the cell membrane that creates the “pore” which allows passage of small molecules (see “A” and “B”). The crystal structure of human Cx26 at 3.5 Å resolution in humans is known,66 and this information was used for the molecular modeling of Cx43. Initially, the amino acid sequence of Cx43 [UniProtKB ID: P08050] was retrieved from UniProtKB database (www.uniprot.org).67 The crystal structure of Cx26 was retrieved from Protein Data Bank (PDB ID: 2ZW3).68 The MODELLER 9v7 program69 was used for sequence alignment and model building.70 The sequence and structural alignment of Cx43 to human Cx26 was carried out using ‘align2d’ program in MODELLER 9v7. This alignment study illustrated 45% sequence identity in the transmembrane region between the two proteins, which suggested that the most important pore region is conserved between Cx26 and Cx43. The produced alignment was then used to generate hemichannel structure with ‘model-multichain’ program in MODELLER 9v7. The quality of the structure was assessed by submitting the modeled structure into PROCHECK.71 The loop building and energy minimization were carried out with Swiss-PdbViewer. The energy computations were done by GROMOS96 implemented in Swiss-PdbViewer. The modeled three dimensional structure of Cx43 was prepared using Schrödinger Suite 2009 Protein Preparation Wizard. All the hydrogens were added to the backbone of the structure and bond orders was assigned appropriately. Exhaustive sampling method was used for the optimization of the hydrogen bonding network, in which the orientation of hydroxyl groups, amide groups of Asn and Gln, and imidazole ring of His residues were optimized. The RMSD (root mean square deviation) of atom displacement for terminating the minimization was set to be 0.30 Å. The minimization was carried out with OPLS2005 force field. The ligand structure of estradiol-17β (CID: 5757), bisphenol A (CID: 6623) (Fig. 3) and adjudin (CID: 9819086) (Fig. 4) were retrieved from NCBI-PubChem database. LigPrep (Version 2.3, Schrödinger, LLC, New York, NY 2009) was used to generate 3D structure with correct chiralities for each ligand used in this study. Grid files represent the volume of the receptor that can be searched for ligand docking. Grid box was set on the centriod of all the residues in the Cx43 and no constraints were selected. The Standard Precision (SP) mode of Glide (Version 5.5, Schrödinger, LLC, New York, NY 2009) was used for the flexible ligand docking. The best binding conformation was selected based on the one having the lowest docking energy from the generated docking solutions. Molecular docking of estradiol-17β into Cx43 generated 32 solutions, in which the best binding conformation having the docking score of −5.40 kcal/mol. The size of binding site surface area of Cx43 was 864.562 Å, and the size of ligand surface was 271.946 Å. These docking studies revealed that side chain oxygen atom of Glu42 (E42, Chain A) makes a hydrogen bond formation with estradiol-17β in the bond length of 1.790 Å. In addition, Gly2 (G2, Chain B), Asp3 (D3, Chain B), Trp4 (W4, Chain A and B), Ser5 (S5, Chain A), Ile31 (I31, Chain A), Ile34 (I34, Chain A), Leu35 (L35, Chain A), Leu37 (L37, Chain A), Gly38 (G38, Chain A) and Val41 (V41, Chain A) were involved in non-bonded interactions such as van der Waals forces. It is important to note that estradiol-17β binds with amino acid residues of N-terminal helix (NTH) and Trans membrane helix 1 (TMH1) of chain A and B alone and it leaves empty space in the pore region. Thus, it is possible that additional units of estradiol-17β may also bind with remaining chains to alter the Cx43 gating mechanism.