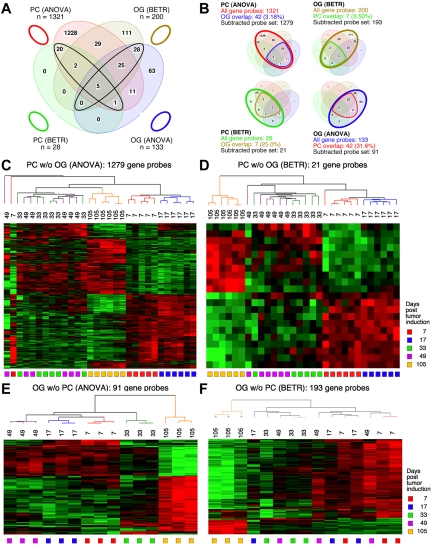
Figure 3.
Gene expression changes during tumor progression in study 2 (validation study). Samples were analyzed as in study 1. Venn diagram showing the number of gene sets found to be significantly variable by ANOVA and BETR analysis of PC (plasma cell) and oil granuloma (OG) samples. Overlap of genes from the different analyses and different samples is indicated. The list of gene sets revealed by ANOVA and BETR analysis of both gene sets, before removal of overlapping genes, can be found in supplemental Tables 5 through 8. (B) Venn diagrams that illustrate the gene subtraction approach used in this study. The ANOVA-revealed PC gene set (top left) consists of 1321 gene probes (red oval), 42 of which also occurred in the ANOVA-revealed OG gene set (blue cone). Thus, the corrected or subtracted ANOVA-PC-gene set consists of 1279 gene probes. The same algorithm applies to the BETR-revealed PC gene set (bottom left), the BETR-revealed OG gene set (top right) and the ANOVA-revealed OG gene set (bottom right). Panels C, D, E, and F present heat maps of changing genes during tumor progression in LMD-isolated PC samples or non-dissected, whole-tissue OG samples after removal of gene sets, as described in panel B. Differently regulated genes identified by either ANOVA or BETR analyses were subjected to unsupervised hierarchical clustering. Up- and down-regulated genes are indicated by red and green, respectively. Time of tumor induction is indicated by colored squares and dendrograms below and above heat maps, respectively.