Abstract
Perfect timing of germination is required to encounter optimal conditions for plant survival and is the result of a complex interaction between molecular processes, seed characteristics, and environmental cues. To detangle these processes, we made use of natural genetic variation present in an Arabidopsis (Arabidopsis thaliana) Bayreuth × Shahdara recombinant inbred line population. For a detailed analysis of the germination response, we characterized rate, uniformity, and maximum germination and discuss the added value of such precise measurements. The effects of after-ripening, stratification, and controlled deterioration as well as the effects of salt, mannitol, heat, cold, and abscisic acid (ABA) with and without cold stratification were analyzed for these germination characteristics. Seed morphology (size and length) of both dry and imbibed seeds was quantified by using image analysis. For the overwhelming amount of data produced in this study, we developed new approaches to perform and visualize high-throughput quantitative trait locus (QTL) analysis. We show correlation of trait data, (shared) QTL positions, and epistatic interactions. The detection of similar loci for different stresses indicates that, often, the molecular processes regulating environmental responses converge into similar pathways. Seven major QTL hotspots were confirmed using a heterogeneous inbred family approach. QTLs colocating with previously reported QTLs and well-characterized mutants are discussed. A new connection between dormancy, ABA, and a cripple mucilage formation due to a naturally occurring mutation in the MUCILAGE-MODIFIED2 gene is proposed, and this is an interesting lead for further research on the regulatory role of ABA in mucilage production and its multiple effects on germination parameters.
Colonizing plants are subject to a wide variety of environmental conditions. For successful adaptation to new habitats, the timing of developmental transitions is especially important. Seed germination is one of these important transitions, as it determines the seasonal environment experienced in further plant life (Huang et al., 2010). Natural populations that develop under distinct environmental conditions may reveal genetic adaptation, which can be used to disentangle the signaling routes that are involved. Seed germination is described by three phases of water uptake. In phase I, the seed imbibes and reinitiates metabolic processes followed by a lag phase (phase II). Further water uptake results in protrusion of the radicle through the testa and endosperm (phase III). The moment of radicle protrusion through the endosperm is considered to be the moment of germination sensu stricto (Finch-Savage and Leubner-Metzger, 2006). To characterize the genetic variation of germination-related traits, we focused on the effect of the environment that a seed perceives during germination rather than the effect of the environment during maternal plant growth, which has been the subject of other studies (Gutterman, 2000; Dechaine et al., 2009; Elwell et al., 2011). Seed content (e.g. oil) is often used as commodity, and modifications to the content therefore can be regarded as seed quality parameters as well. To prevent confusion, we use the term “seed performance” to indicate that the focus of our study is restricted to seed germination characteristics.
The production of high-quality crop seed not only entails knowledge about maternal plant growth, harvesting, and storage of seeds but also of germination conditions (Rivero-Lepinckas et al., 2006). To obtain better germination and field performance, many seed companies rely on enhancement methods, such as seed priming and coating and/or pelleting, but these methods are reaching their limits. Dissecting the molecular mechanisms underlying seed germination and its tolerance to the environment may unlock the full genetic potential and enable targeted breeding for seed performance.
In this study, we used a recombinant inbred line (RIL) population derived from two Arabidopsis (Arabidopsis thaliana) ecotypes: Bayreuth (Bay-0), which originates from a fallow land habitat in Germany, and Shahdara (Sha), which grows at high altitude in the Pamiro-Alay mountains in Tadjikistan (Loudet et al., 2002). The Bay-0 × Sha RIL population has been used in many previous studies to map quantitative trait locus (QTL) positions for root morphology (Loudet et al., 2005; Reymond et al., 2006), anion content (Loudet et al., 2003a), nitrogen use efficiency (Loudet et al., 2003b), cell wall digestibility (Barriere et al., 2005), carbohydrate content (Calenge et al., 2006), sulfate content (Loudet et al., 2007), leaf senescence (Diaz et al., 2006), morning-specific growth (Loudet et al., 2008), and cold-dark germination (Meng et al., 2008). We have used the natural variation present in this RIL population to map the response of germination characteristics to environmental conditions to which a seed is exposed.
Freshly harvested viable Arabidopsis seeds often do not germinate even when placed under conditions favorable for germination. This event, called primary dormancy, is shown to be subject to natural variation (Bentsink et al., 2010). In many Arabidopsis ecotypes, this primary dormancy is released after a period of dry storage at room temperature. Another dormancy-breaking treatment is cold stratification, where seeds are imbibed in water and stored at 4°C in the dark for 4 d before putting them into optimal conditions for germination (Finch-Savage and Leubner-Metzger, 2006). Unfavorable conditions during seed germination may result in a changed rate or even failure of germination. In Arabidopsis, it has been shown that the responsiveness to temperature is closely related to the level of after-ripening (Tamura et al., 2006). High salt concentrations induce osmotic stress and ion toxicity, resulting in both a delay and reduction of maximum germination (Galpaz and Reymond, 2010). Often, these different environmental stresses are interconnected and will cause osmotic and associated oxidative stresses (Zhu, 2002; Chinnusamy et al., 2004). The plant hormone abscisic acid (ABA) plays a predominant role in plant responses to different environmental stresses and can activate various signal transduction pathways leading to a global change in transcription (Finkelstein et al., 2002; Xiong et al., 2002). Exogenous application of ABA during germination results in a distinction between testa and endosperm rupture. At certain concentrations, the testa will rupture but germination sensu stricto (radicle protrusion through the endosperm) will be inhibited. This phenomenon, caused by reduced weakening of the endosperm cap, is the consequence of a complex interplay between ABA, GA, and ethylene signals (Linkies et al., 2009). In this report, we determined germination sensu stricto for primary dormancy in freshly harvested seeds, germination of fully after-ripened seeds with and without a preceding cold stratification period (for conditions, see “Materials and Methods”), and germination under various stress conditions (low/high temperature, salt/osmotic stress, and ABA) to assess natural variation in the Bay-0 × Sha RIL population. Additionally, seed morphology (size and length) and flowering time were phenotyped, as they have been shown to be strong determinants of plant trait variation (Chiang et al., 2009; Orsi and Tanksley, 2009; Elwell et al., 2011). We correlated these traits to our germination-related traits to evaluate possible causality. In total, this analysis resulted in 327 trait scores over different harvests. Evaluation of these high numbers of phenotypes demanded methods of QTL analysis that extended beyond mapping of individual traits and that allowed comprehensive and comprehensible visualization.
Analysis of natural variation that is captured in well-defined recombinant inbred populations has been shown to be a powerful tool to detect important loci that influence the traits under study (Alonso-Blanco et al., 2009). To uncover the loci with genetic variation, a statistical framework is needed. For this, any programming language can be used that supports statistics. In the life sciences, the statistical language R is often the prime candidate. R is open source, contains the latest in statistical analysis methods, and has a large community for help and support (http://www.r-project.org/). Furthermore, it has the R/qtl package (Broman et al., 2003), which contains an array of different QTL-mapping methods, including Single Marker Mapping, Interval Mapping, and Multiple QTL Mapping (MQM; Arends et al., 2010). Although all possibilities to perform a detailed QTL analysis, including data preprocessing and output formatting, are present in R, it requires extensive knowledge of the R syntax to combine all necessary steps in a single analysis protocol that can loop through hundreds or thousands of traits. In this paper, we present a script that can perform these tasks. This type of automated analysis combined with efficient data visualization is a necessary step to keep up with the increasing rate of biological data production. For using single-trait mapping, the effect of a certain treatment (e.g. germination at high temperature) must be corrected by the germination characteristics under control conditions. Here, we subtracted the observed germination under stress conditions from values for germination under control conditions. This correction can lead to complicated interpretation, especially when the environment under study affects loci with already strong effects under control conditions. Furthermore, it can reduce statistical power, due to summation of the error components. Therefore, we performed an additional analysis using a QTL-by-environment (QTL×E) approach (Jansen et al., 1994; Malosetti et al., 2004; Moreau et al., 2004; Eeuwijk et al., 2006). Instead of considering individual responses, one can then treat the stress conditions as a set of environmental perturbations and evaluate a single trait (such as germination percentage). Because several environments are taken into account simultaneously, the statistical power to detect loci that are affected by several environments increases, and interpretation becomes more intuitive as the need for correcting the stress response by the control response is eliminated (Boer et al., 2007; Payne et al., 2011).
The Bay-0 × Sha RIL population consists of 420 lines that were genotyped in the F6. This relatively low degree of inbreeding provoked residual heterozygosity present at almost all genome positions. This residual heterozygosity can be used to confirm QTL positions, as it provides a possibility to study both parental alleles at the locus of interest in an elsewhere homozygous background (Tuinstra et al., 1997). In contrast to conventional near isogenic lines, the genetic background of heterogeneous inbred lines (heterogeneous inbred families [HIFs]) consists of a mix of the two parental genomes. The availability of a genome-wide set of HIF lines for the Bay-0 × Sha RIL population provides fast and accurate means to confirm QTL loci.
RESULTS
Phenotyping Seed Germination
To map the genetic architecture of seed germination traits, we have used the core population (165 lines) of the Bay-0 × Sha RIL population (Loudet et al., 2002). Flowering time was recorded during maternal plant growth and showed good correlation (Pearson r2 = 0.75) with previously published data of this population (Loudet et al., 2002). We used freshly harvested seeds (2 weeks of after-ripening) and tested germination with and without stratification (for conditions, see “Materials and Methods”). Furthermore, we tested germination of fully after-ripened seeds and assayed germination under several stress conditions, with and without stratification (Table I). Germination was measured with the automated scoring system, the Germinator (Joosen et al., 2010). Using this package, we were able to describe the cumulative germination curve under all conditions tested. The germination curve can accurately be described by extracting five parameters: Gmax = maximum germination; U8416 = uniformity of germination, time between 16% and 84% germination; t10 = initiation of germination, time to reach 10% germination; t50 = rate of germination, time to reach 50% germination; and AUC = area under the germination curve until 100 h.
Table I. Overview of traits in this study and the harvest(s) used for the measurements.
The indicated color code is used in all figures throughout this paper. For each mentioned experiment, Gmax, AUC, t50, t10, and U8416 were determined. Abbreviations in codes are as follows: AR, after-ripened; CD, controlled deterioration; NS, no stratification; WS, with stratification; Δ, difference.
To reduce environmental variation, we took great care in the growth and harvest of the maternal plants. We used a fully randomized setup and grew the population twice in a climate chamber. In the first growth, we separated the harvest into three blocks (ABC), each with three to five plants per RIL. In the second growth, we pooled the harvest of four to seven plants per RIL (D). The overall Pearson correlation between blocks A-B, A-C, and B-C was higher compared with A-D, B-D, and C-D (Supplemental Table S1). Broad-sense heritability was calculated with the QTL data analysis tools in Genstat 14, using the preliminary single environment analysis and adding the block as an additional fixed term (Table II). Heritability values can range between 0 (no heritability) and 1 (maximum heritability). Overall, heritability was high, indicating a large genetic variance and a small effect of the different harvests. The lower heritability for maximum germination at low temperature (AR.NS.Cold) and after cold stratification (AR.WS) is the result of low genetic variance for these traits, as many of the lines germinated to 100% under these conditions. However, we were able to capture the genetic variation for these traits in the other parameters (AUC, t50, t10, and U8416). Although this breadth of phenotypic screens is common today, easy tools for performing high-throughput QTL mapping and generating clear overview figures were not available. It is important to detect and correct data errors, enable selection of traits that should be analyzed in more detail, detect possible epistatic interactions, and find strong correlations between phenotypes. Because these are crucial steps in determining biological relevance, we invested in the development of an automated analytical protocol that allowed large-scale single-trait QTL analysis.
Table II. Overview of the broad-sense heritability scores.
Included are those traits for which different blocks were tested (trait code descriptions can be found in Table I).
| Trait | Gmax | AUC | t50 | t10 | U8416 |
| AR.NS | 0.82 | 0.87 | 0.86 | 0.79 | 0.82 |
| AR.NS.Cold | 0.51 | 0.77 | 0.73 | 0.66 | 0.48 |
| AR.NS.Mannitol | 0.61 | 0.79 | 0.70 | 0.55 | 0.62 |
| AR.NS.NaCl | 0.90 | 0.94 | 0.80 | 0.76 | 0.43 |
| AR.WS | 0.63 | 0.79 | 0.78 | 0.72 | 0.72 |
| AR.WS.NaCl | 0.91 | 0.93 | 0.86 | 0.78 | 0.70 |
| Fresh.NS | 0.92 | 0.94 | 0.81 | 0.70 | 0.76 |
| Fresh.WS | 0.40 | 0.81 | 0.87 | 0.84 | 0.76 |
Single-Trait QTL Mapping
To evaluate the response of germination to a certain treatment, we first subtracted the observed germination at test conditions from germination at the proper control conditions. For example, the effect of NaCl on germination after cold stratification is determined by subtracting Gmax on NaCl from Gmax on water. This subtraction was reversed for the rate and uniformity parameters to correct the reversed nature of these parameters (e.g. slower germination results in a larger t10 and t50). Table I gives an overview of all corrections that have been applied.
An analytical protocol was designed, using the popular R/qtl package of R, to analyze trait data of recombinant inbred populations with the multiple-QTL-model approach (Arends et al., 2010). When performing a detailed QTL analysis, it is important that several steps are performed or checked. Missing genotypic data are imputed, and a recombination frequency plot is generated (Fig. 1A). In the next step, the quality of the trait data is investigated. Outliers are detected and removed using a Z-score transformation with a user-defined threshold. To estimate the effect of data normalization on MQM mapping, we have used the distribution analyzer version 1.2 (http://www.variation.com). A logarithm of the odds (LOD) score correlation plot comparing raw and normalized data (Supplemental Fig. S1) clearly shows that this does not affect the output. Therefore, we decided to use nontransformed data instead of fitting a polynomial distribution without a proper biological rationale. As an extra control, the results of the MQM mapping were always compared with standard interval mapping, using the parametric model with Haley-Knott regression (Haley and Knott, 1992; Fig. 1B). The whole-genome additive effect was estimated based on the nontransformed data as half the difference between the phenotypic averages for the two homozygotes (Fig. 1C).
Figure 1.
R/qtl output for the effect of 100 mm NaCl on the maximum germination without stratification. A, Pairwise recombination fractions. B, LOD profile comparison between MQM and Haley-Knott (scan.one) interval mapping. C, Genome-wide additive effect based on raw phenotype data. D, Genetic map showing the significant QTL markers. E, Interaction plot showing the effect-size comparison between markers MSAT305754 and MSAT518662 at the Sha (AA) and Bay-0 (BB) alleles. F, Circle plot showing epistatic interactions between all significant QTLs.
R/qtl MQM uses a backward elimination of cofactors. As a rule of thumb, one can select a maximum of n-16 initial cofactors with this procedure (Jansen, 2008), with n being the number of lines in the RIL population. In our script, a cofactor file can be provided with the selection of the initial cofactors. When no cofactor file is provided, the analysis will be performed without cofactors, resulting in an analysis comparable to the composite interval-mapping method. For the analysis of the Bay-0 × Sha population, we selected 39 out of 69 markers as possible cofactors. Cofactors were selected based on their quality (least amount of missing data or heterozygous status) and physical position in centimorgan (cM), attempting to obtain intervals of about 10 cM. Although the procedure allows the selection of all 69 markers as cofactors, this does not improve mapping and only lowers statistical power due to the multiple testing correction in the permutation analysis. The provided cofactor file is used to perform automated backward elimination of cofactors. Backward elimination is performed to remove cofactors that do not significantly contribute to the fit of the initial model. This is achieved by comparing Akaike’s information criteria of the different models (Jansen, 1993). Using the final selected QTL model, the mapping LOD scores are calculated for all genetic markers. Plots showing all significant markers are produced automatically (Fig. 1D). We have used the procedure described to map all 327 individual measurements (Supplemental Table S2), but to enhance the readability of this paper, we only show average values for each trait (94 traits; Supplemental Table S3).
Detection of Epistatic Interactions
Detection of epistatic interactions with the relatively limited sample sizes that are common in RIL populations is often cumbersome (Li et al., 2010). However, a useful hint of epistasis can be obtained when clear effects are visible between loci, and the same interaction can be detected when measuring multiple traits. We calculated epistatic interactions between the QTL loci that are detected with the multiple QTL models from MQM. All possible combinations of the detected QTL loci were used to calculate the estimated interaction effect (Fig. 1E). Interactions will be reported when their biological effect size is above a user-defined threshold, which is based on the number of sd differences between two interacting loci. In this study, we have set this threshold to 8. For each single trait, a graphical visualization shows all interactions, using the circle-plot routine from R/qtl (Fig. 1F).
Multiple Trait Visualizations
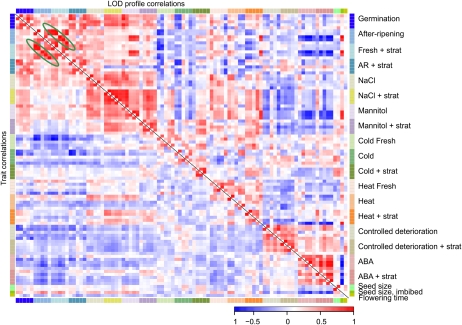
The interpretation of large numbers of phenotypes requires comprehensive visualization methods. Therefore, we produced several outputs that can help to dissect and interpret the data. A correlation plot based on trait values enabled a quick overview of similarities between all input traits (Fig. 2, bottom left panel). After QTL mapping of all traits, this was also done based on the LOD profiles (Fig. 2, top right panel). This plot shows a strong correlation between the effect of after-ripening and stratification on fresh seeds, indicating that in the Bay-0 and Sha ecotypes, both dormancy-breaking treatments resulted in total recovery of germination. All other stress treatments had a negative effect on germination and resulted in negative trait correlations, as compared with after-ripening and stratification. Interestingly, the same structure was observed when studying the LOD profile correlations. This indicates that most of the variation is well captured in the genetic analysis. Neither dry seed size nor flowering time correlated significantly with any of the germination parameters. Imbibed seed size appeared to have a negative correlation with germination in the presence of ABA and a positive correlation with the rate of germination. This correlation was strongest at LOD-profile levels.
Figure 2.
Correlation between all trait values (bottom left panel) and between all LOD profiles (top right panel). Five parameters (Gmax, AUC, t50, t10, and U8416) per experiment are shown. A precise description of the trait values can be found in Table I. Green circles indicate an example of the close correlation between “after-ripened” and “fresh-with-stratification” trait values.
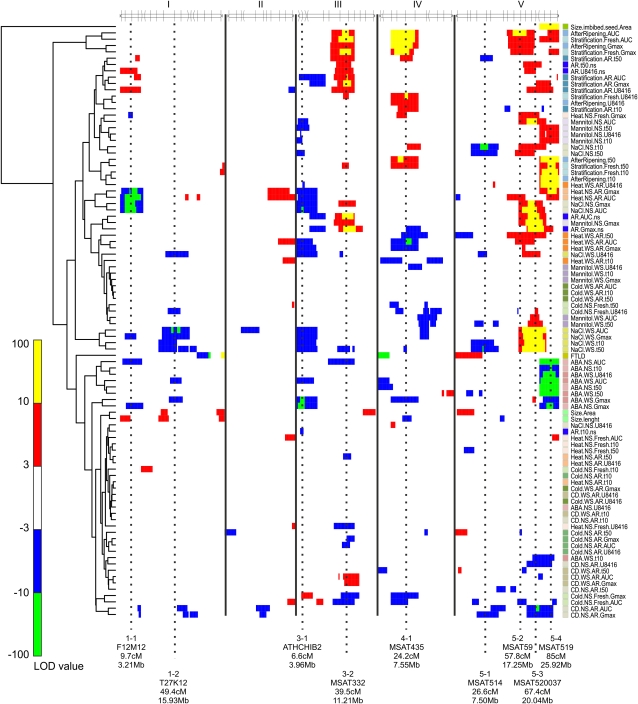
Furthermore, the analytical script creates heat maps of all LOD scores. These heat maps can be used to interpret the genetic landscape of the studied traits. A heat map of all LOD scores clustered by traits (one-way) with Hclust (Murtagh, 1985) is created to visualize similarities among different traits (Fig. 3). According to a procedure described by Breitling et al. (2008), the heat map shows several significant hotspots in the genome (Fig. 3). These appear to control different traits and confirm the high level of interconnection among the responses to different environmental stresses, which is in agreement with earlier findings of pervasive genetic buffering (Fu et al., 2009). They found that only a few influential hotspot regions cause major phenotypic variation across a range of environmental conditions, whereas the largest fraction of molecular variants is silent at the phenotypic level. Furthermore, a clear separation in the clustering can be observed between dormancy/mannitol/salt/heat/cold QTLs compared with ABA and germination after controlled deterioration QTLs. This is mainly caused by the large QTL with a reversed effect on the bottom of chromosome V.
Figure 3.
A clustered heat map showing the LOD profiles of the measured traits. Columns indicate chromosome position along the five chromosomes; rows indicate individual-trait LOD profiles. A false-color scale is used to indicate the QTL significance. Positive values (yellow and red) represent a larger effect of the treatment in Sha, and negative values (blue and green) represent a larger effect of the treatment in Bay-0. Dashed lines indicate major QTL positions, which are further discussed in Table IV. Clustering on the left shows correlation between QTL profiles.
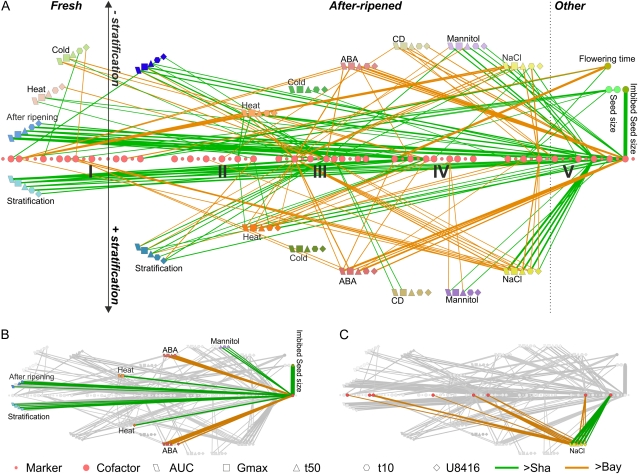
The protocol stores all LOD profiles in a single tab-delimited text file, allowing further analysis in most available statistical and/or microarray analysis software suites. A second output file summarizes all QTL results, providing an overview of all detected QTLs, their LOD scores, positions, confidence intervals, and directions (Supplemental Table S4 shows all 327 traits, Supplemental Table S5 shows the output for the average traits described here). An adjusted result file in the .sif format allows direct import into Cytoscape. Using Cytoscape, we created a marker-trait network of QTL positions, with nodes indicating markers or traits and edges representing LOD scores and directions, allowing an alternative method to visualize many trait QTLs in one figure (Fig. 4A; Supplemental File S1). One advantage of loading a QTL network in Cytoscape is the dynamic nature of the program, which allows ordering, filtering, and selection in all directions. Figure 4B shows an example where a specific marker (MSAT519) was selected to show all traits that have a significant QTL at this locus, visualized by the adjacent edges and connected nodes. In Figure 4C, we selected a single trait (germination on NaCl with stratification) to show all significant QTL positions for this trait.
Figure 4.
Cytoscape marker-trait network. A, Significant QTL positions are indicated by a connection between traits and markers. Edge colors indicate the direction of the QTL effect, and line width indicates the LOD score. B, Subnetwork showing all traits with a significant QTL at marker MSAT519. C, Subnetwork showing all markers with significant QTLs for germination on NaCl with stratification.
QTL×QTL Interactions
Epistatic interactions between QTLs can help to elucidate meaningful colocalizations and will enable an efficient design of follow-up experiments. Besides the visualization of the epistatic interactions per trait (Fig. 1F), our script creates an output that can help to visualize all detected epistatic interactions in a single plot. This output file in .sif format summarizes all detected epistatic interactions (Fig. 5; Supplemental File S2). Among others, clear hotspots of epistatic interactions between QTL loci on chromosomes III, IV, and V (ATHCHIB2 + MSAT332, MSAT435, and MSAT520037 + MSAT519, respectively) were observed for germination on salt (yellow lines) and dormancy (blue lines). Next to the importance of detecting possible interacting loci, this QTL×QTL analysis provides additional arguments for colocating QTLs to be of similar genetic origin. Overall, the creation of this type of summarizing figure greatly facilitates the interpretation of large data sets.
Figure 5.
Epistatic interaction network. Nodes indicate markers (small circles) and selected cofactors (large circles). Edges represent the detected significant epistatic interactions (edge colors represent traits). A precise description of the trait values can be found in Table I.
QTL×E Interaction
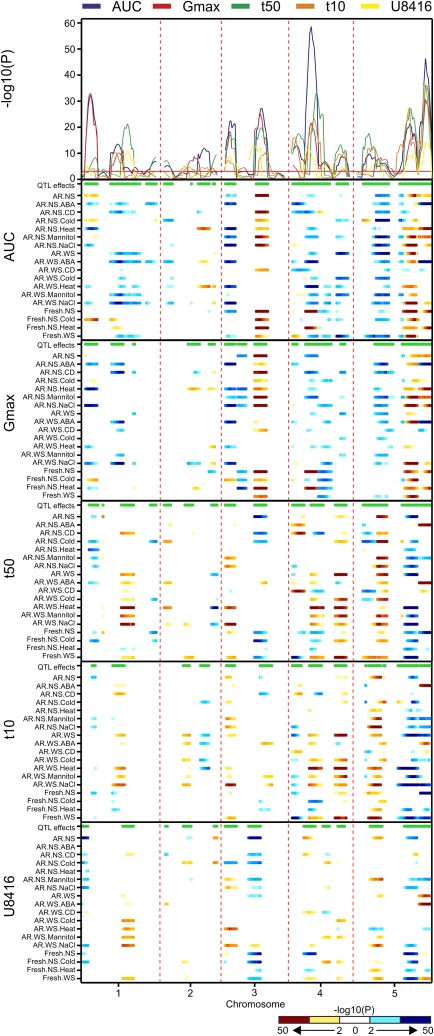
To obtain a parameter for the response, we had to correct all values with their proper control condition values. This sometimes led to complex interpretation, which can be circumvented by using the noncorrected germination parameters and modeling them over the various environmental conditions that were tested. Because several environments are taken into account simultaneously, the statistical power to detect loci that are affected by several environments increases, and interpretation becomes more intuitive as the need for correcting the stress response by the control response is eliminated. By using this approach, the sensitivity of a specific QTL for environmental conditions can be determined for each separate germination parameter. Details about the procedure are described in “Materials and Methods.” Results are summarized in Figure 6. The final model P value profiles (Fig. 6, top panel) clearly show the great consistency between the five germination parameters that we measured. However, a closer look also reveals loci that are affecting different germination curve parameters. For example, the QTL on the top of chromosome V is not detected by measuring maximum germination but is well defined when using t50 or t10 as parameter. As expected, the parameter AUC outperforms the others, as it represents a combined value for maximum germination percentage, rate, and uniformity. For comparison of the environment-specific QTL effects for the five different germination parameters (Fig. 6, five lower panels), the effects could be compared with germination under control conditions. For example, after-ripened seeds without stratification (AR.NS) can serve as a reference for the stress treatments (AR.NS.ABA, AR.NS.CD, AR.NS.Cold, AR.NS.Heat, AR.NS.Mannitol, AR.NS.NaCl). The same analogy holds true for after-ripened seeds without stratification (AR.NS) and freshly harvested seeds without stratification (Fresh.NS). In this way, stress-specific QTLs on chromosome II and the top of chromosome III can easily be identified. Interestingly, some QTLs, including germination at low temperature (top of chromosome I) and germination in the presence of exogenous ABA (bottom of chromosome V), displayed opposite effects on germination when compared with the other treatments. In Table III, the environment-specific effect sizes are summarized for the major loci. A complete overview of effect sizes and explained variances for all detected loci can be found in Supplemental Table S6.
Figure 6.
Genome scan for QTL×E effects for seed germination. The P values for the main effects of the different germination parameters are shown in the top panel. The red horizontal line is the genome-wide significance threshold. The five lower panels show the environment-specific QTL effects. The horizontal green bar at the top of each panel indicates significant environment-specific effects. For both Gmax and AUC, a larger effect of the Sha allele is indicated in yellow-red (Bay-0 in cyan-blue). The color scale is opposite for the t50, t10, and U8416 parameters due to the inversed nature of these parameters.
Table III. Significant environment-specific QTL effects (P < 0.05).
Positive values for AUC and Gmax indicate a larger effect of the Sha allele, and negative values for AUC and Gmax indicate a larger effect of the Bay-0 allele. This is opposite for t50, t10, and U8416 values due to the inversed nature of these parameters. Environments are as mentioned in Table I, and loci are as indicated in Figure 3.
| Locus | 1-1 |
1-2 |
3-1 |
3-2 |
4-1 |
5-1 |
5-2 |
5-3 |
||||||||||||||||||||||||||||||||
| cM | 9 |
50 |
5 |
44 |
24 |
14 |
63 |
83 |
||||||||||||||||||||||||||||||||
| Environment | AUC | Gmax | t50 | t10 | U8416 | AUC | Gmax | t50 | t10 | U8416 | AUC | Gmax | t50 | t10 | U8416 | AUC | Gmax | t50 | t10 | U8416 | AUC | Gmax | t50 | t10 | U8416 | AUC | Gmax | t50 | t10 | U8416 | AUC | Gmax | t50 | t10 | U8416 | AUC | Gmax | t50 | t10 | U8416 |
| % | h | % | h | % | h | % | h | % | h | % | h | % | h | % | h | |||||||||||||||||||||||||
| AR.NS | 1.3 | −0.8 | −2.0 | −1.3 | 0.8 | 3.5 | 4.6 | −1.8 | −2.5 | −1.2 | −3.1 | 1.4 | 1.8 | 1.8 | −2.0 | −1.4 | −1.6 | 2.8 | ||||||||||||||||||||||
| AR.NS.ABA | −1.6 | −5.8 | −1.4 | −6.1 | 5.7 | −2.5 | −9.9 | 4.6 | 5.2 | −4.6 | −12.1 | 23.2 | 15.7 | 14.2 | ||||||||||||||||||||||||||
| AR.NS.CD | −1.5 | −3.2 | −6.8 | 2.5 | 3.1 | 1.4 | 5.3 | 9.0 | −3.5 | −2.1 | −2.1 | −2.9 | −7.8 | −3.5 | −2.2 | 1.9 | −2.8 | |||||||||||||||||||||||
| AR.NS.Cold | 0.9 | −1.3 | −1.3 | 1.2 | −1.3 | 1.3 | 1.4 | −1.6 | −1.4 | −1.3 | 1.0 | −2.2 | −1.3 | |||||||||||||||||||||||||||
| AR.NS.Heat | −3.5 | −7.4 | −1.8 | −4.3 | −5.8 | 2.3 | 6.1 | −2.2 | −4.2 | 2.8 | 5.0 | −1.8 | 5.2 | 8.6 | ||||||||||||||||||||||||||
| AR.NS.Mannitol | −1.9 | −1.9 | −2.6 | −2.9 | 2.5 | 1.8 | 2.2 | 3.4 | 11.7 | −2.7 | −2.0 | −6.1 | 3.1 | 6.7 | −3.3 | −2.0 | 3.0 | 5.8 | −2.3 | −1.6 | −3.4 | |||||||||||||||||||
| AR.NS.NaCl | −2.1 | −7.8 | −1.8 | −2.0 | −4.2 | −6.8 | 2.5 | 2.0 | 1.7 | 3.9 | 10.4 | −1.3 | 4.5 | 8.6 | −3.6 | −2.1 | 1.9 | 5.3 | −1.7 | |||||||||||||||||||||
| AR.WS. | 0.4 | −0.8 | 0.6 | 0.5 | 0.4 | −0.6 | −0.7 | −0.4 | 0.5 | 0.5 | 1.2 | 0.4 | −1.3 | −0.4 | −0.7 | 1.2 | ||||||||||||||||||||||||
| AR.WS.ABA | −2.1 | −1.9 | −4.8 | −4.4 | −7.2 | 2.7 | 6.0 | −5.2 | −8.7 | 2.5 | 1.9 | −5.3 | 2.4 | 1.5 | 3.8 | 8.3 | −12.2 | −14.0 | 10.8 | 2.4 | 17.8 | |||||||||||||||||||
| AR.WS.CD | −1.5 | −4.5 | 1.3 | 4.2 | 5.9 | −4.8 | −1.9 | 4.2 | −2.7 | 1.8 | 1.4 | |||||||||||||||||||||||||||||
| AR.WS.Cold | 1.1 | 1.7 | −1.2 | 1.3 | 1.2 | −1.4 | ||||||||||||||||||||||||||||||||||
| AR.WS.Heat | −1.6 | −2.4 | 1.6 | 1.0 | 1.2 | −3.5 | −2.9 | 1.1 | 0.6 | 1.2 | −4.6 | −2.7 | 1.6 | 1.4 | 3.3 | 2.6 | −2.2 | −1.1 | −0.8 | |||||||||||||||||||||
| AR.WS Mannitol | −2.6 | −2.3 | 1.7 | 1.3 | 1.6 | 0.8 | 0.9 | −2.1 | 1.6 | 0.9 | 2.0 | −2.2 | ||||||||||||||||||||||||||||
| AR.WS.NaCl | −1.9 | −4.4 | 0.9 | −1.7 | −4.6 | −6.4 | 2.7 | 1.8 | 2.9 | −4.8 | −5.7 | 2.8 | 1.6 | 2.4 | 0.7 | 1.6 | −3.1 | 1.8 | 1.3 | 6.5 | 9.1 | −3.8 | −1.5 | −2.3 | 2.5 | 3.4 | −1.4 | |||||||||||||
| Fresh.NS | −2.6 | −3.0 | 1.8 | 1.2 | 2.4 | 7.5 | 15.3 | −2.4 | −5.9 | 8.5 | 15.8 | −2.8 | −6.7 | 5.9 | 7.5 | −3.0 | −7.2 | 3.7 | 6.2 | −4.9 | −5.2 | 3.1 | ||||||||||||||||||
| Fresh.NS.Cold | 2.1 | 3.7 | −4.2 | −4.3 | 1.5 | −2.1 | 4.5 | 2.8 | 5.1 | −5.3 | −7.3 | −3.5 | −4.2 | 2.2 | 6.5 | −4.6 | −5.5 | |||||||||||||||||||||||
| Fresh.NS.Heat | −4.6 | 2.8 | −3.2 | 6.5 | 18.0 | −3.2 | −0.8 | 5.7 | 16.1 | −3.6 | −1.0 | 4.2 | 10.0 | −6.6 | −4.0 | −1.2 | 5.2 | 9.0 | −4.4 | −1.0 | ||||||||||||||||||||
| Fresh.WS | 0.6 | −0.7 | 0.5 | 0.6 | −0.8 | 0.6 | 0.5 | 0.9 | 0.9 | −0.6 | −0.6 | −0.9 | 0.9 | 0.7 | 0.6 | −1.2 | 0.9 | 2.2 | 1.0 | −1.8 | −0.6 | −1.0 | −0.9 | 0.8 | ||||||||||||||||
QTL Confirmation
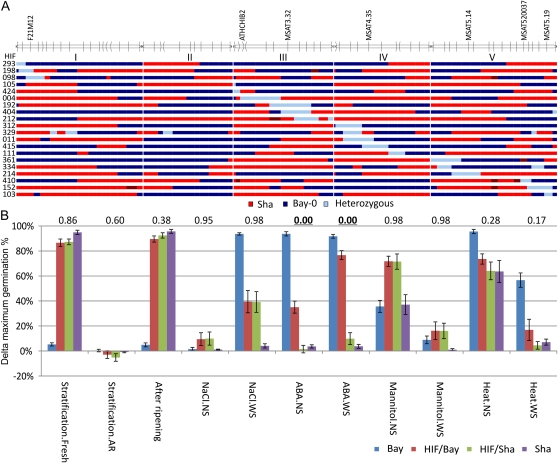
Taking advantage of the residual heterozygosity present in the F6 generation of the Bay-0 × Sha population, combined with the large population size, we were able to confirm several QTLs following the HIF approach. In short, RILs that are heterozygous at the locus of interest were selected in the next generation for lines homozygous for both parental alleles. These “families” are near isogenic lines that can be used to confirm the observed allelic effects (Fig. 7A). We applied this strategy for seven of the major QTLs that we detected in this study and tested the five germination parameters for 11 different conditions. For a single parameter (Gmax) and a single HIF (line HIF103), the analytical procedure is summarized in Figure 7B. Traits that could be confirmed by one or several HIF lines are indicated in Table IV. An overview of all HIF results can be found in Supplemental Table S7.
Figure 7.
Confirmation of QTLs with a HIF approach. A, The blue/red bars indicate the allelic distribution of the HIF lines used (blue = Bay-0, red = Sha, light blue = segregating). The five chromosomes are indicated at the top with the nearest genetic marker for the seven major loci. B, Example analysis for HIF103 (segregating at MSAT5.19, at the bottom of chromosome V). Indicated is the response for maximum germination for 11 conditions compared with the control. Error bars represent se of at least six replicates. Responses are calculated by subtracting the test sample from the control sample, as indicated in Table I. Numbers above the graph show the t test significance for the responses as measured between HIF/Bay-0 versus HIF/Sha (significant values [P < 0.05] are in boldface).
Table IV. Colocations at the major QTL loci.
| QTLa | Trait | HIF Confirmationb | Marker | LOD | cM/Mb | Interval | Effect | Colocalization with Other Seed Studies |
| cM | ||||||||
| 1-1 | NaCl.NS.Gmax | F21M12 | 13.1 | 9.7/3.21 | 5–13 | >Bay-0 | Cloned QTL for salt and ABA sensitivity during germination and seedling growth in Ler × Sha, RAS1 (Ren et al., 2010) | |
| NaCl.NS.AUC | 14.1 | 6–13 | >Bay-0 | |||||
| NaCl.WS.Gmax | 098/198 | 4.6 | 4–14 | >Bay-0 | ||||
| ABA.NS.Gmax | 098 | 4.7 | 5–22 | >Bay-0 | ||||
| ABA.NS.AUC | 098 | 6.6 | 4–14 | >Bay-0 | ||||
| Heat.NS.AR.Gmax | 098 | 10.1 | 6–18 | >Bay-0 | QTL for germination after heat shock in Ler × Sha (Clerkx et al., 2004) | |||
| Heat.NS.AR.AUC | 098 | 14.4 | 6–18 | >Bay-0 | ||||
| 1-2 | NaCl.WS.Gmax | T27K12 | 7.9 | 49.1/15.93 | 42–54.7 | >Bay-0 | QTL for germination on salt in Ler × Sha (Galpaz and Reymond, 2010) | |
| NaCl.WS.U8416 | 3.6 | 38–62 | >Bay-0 | |||||
| NaCl.WS.AUC | 9.2 | 41–57 | >Bay-0 | |||||
| ABA.WS.Gmax | 4.9 | 44–54.7 | >Bay-0 | |||||
| 3-1 | Stratification.AR.AUC | ATHCHIB2 | 3.4 | 6.6/3.96 | 0–19 | >Bay-0 | QTL for germination on ABA and salt in Ler × Sha (Clerkx et al., 2004) | |
| NaCl.NS.Gmax | 6.0 | 2–16 | >Bay-0 | |||||
| NaCl.NS.t50 | 4.4 | 0–8 | >Bay-0 | QTL for germination on polyethylene glycol in Bay-0 × Sha, OSMOTIC SENSITIVITY RESPONSE2 (Vallejo et al., 2010) | ||||
| NaCl.NS.AUC | 9.7 | 1–7.9 | >Bay-0 | |||||
| NaCl.WS.Gmax | 424 | 5.9 | 1–16 | >Bay-0 | ||||
| NaCl.WS.t50 | 004 | 7.9 | 0–14 | >Bay-0 | ||||
| NaCl.WS.U8416 | 004 | 4.0 | 0–19 | >Bay-0 | ||||
| NaCl.WS.t10 | 424 | 4.9 | 0–17 | >Bay-0 | ||||
| NaCl.WS.AUC | 424 | 9.6 | 1–14 | >Bay-0 | ||||
| Mannitol.NS.AUC | 3.6 | 0–16 | >Bay-0 | |||||
| ABA.NS.Gmax | 424/004 | 10.7 | 1–7.9 | >Bay-0 | ||||
| ABA.WS.Gmax | 105/424/004 | 10.8 | 2–7.9 | >Bay-0 | ||||
| ABA.WS.AUC | 424/105 | 6.5 | 0–7.9 | >Bay-0 | ||||
| Cold.NS.Fresh.Gmax | 3.4 | 0–19 | >Sha | |||||
| Heat.NS.AR.Gmax | 424 | 7.6 | 3.2–7.9 | >Bay-0 | ||||
| Heat.NS.AR.AUC | 6.9 | 3.2–19 | >Bay-0 | |||||
| Heat.WS.AR.Gmax | 4.7 | 0–17 | >Bay-0 | |||||
| Heat.WS.AR.U8416 | 424 | 3.4 | 4–17 | >Bay-0 | ||||
| Heat.WS.AR.AUC | 5.0 | 1–13 | >Bay-0 | |||||
| 3-2 | Stratification.Fresh.Gmax | 192/404 | MSAT332 | 11.3 | 39.5/11.21 | 36–50 | >Sha | Dormancy QTL; delay of germination, DOG6 (Bentsink et al., 2010) |
| Stratification.Fresh.AUC | 004/192/404 | 12.9 | 37–50 | >Sha | ||||
| Stratification.AR.Gmax | 004/192 | 8.0 | 35–52 | >Sha | ||||
| Stratification.AR.t50 | 404 | 6.7 | 33–48 | >Sha | ||||
| Stratification.AR.U8416 | 8.1 | 35–48 | >Sha | QTL for dormancy and germination on salt in Ler × Sha (Clerkx et al., 2004) | ||||
| Stratification.AR.AUC | 404 | 7.8 | 36–48 | >Sha | ||||
| AfterRipening.Gmax | 004/192/404 | 9.0 | 35–48 | >Sha | ||||
| AfterRipening.AUC | 404 | 10.0 | 36–48 | >Sha | ||||
| NaCl.WS.t50 | 004 | 3.8 | 33–52 | >Bay-0 | QTL for cold-dark germination in Bay-0 × Sha, COLD-TOLERANT DARK GERMINATION-3 (CDG-3;Meng et al., 2008) | |||
| Mannitol.NS.Gmax | 192/404 | 6.7 | 36–52 | >Sha | ||||
| ABA.NS.AUC | 8.4 | 32–46 | >Bay-0 | |||||
| Cold.NS.Fresh.AUC | 8.0 | 36–52 | >Bay-0 | |||||
| Heat.NS.Fresh.U8416 | 4.8 | 33–48 | >Bay-0 | |||||
| 4-1 | Stratification.Fresh.Gmax | 329/415 | MSAT435 | 14.6 | 24.2/7.55 | 16–26 | >Sha | Dormancy QTL; delay of germination, DOG18 (Bentsink et al., 2010) |
| Stratification.Fresh.t50 | 111 | 8.4 | 17–32 | >Sha | ||||
| Stratification.Fresh.U8416 | 9.9 | 17–32 | >Sha | |||||
| Stratification.Fresh.AUC | 329/415 | 28.8 | 17–26 | >Sha | ||||
| Stratification.AR.t50 | 4.0 | 19–33 | >Sha | |||||
| Stratification.AR.t10 | 011 | 5.7 | 20–37 | >Sha | ||||
| AfterRipening.Gmax | 329/011/415/111 | 20.4 | 16–26 | >Sha | ||||
| AfterRipening.t50 | 9.9 | 16–28 | >Sha | |||||
| AfterRipening.U8416 | 8.8 | 17–31 | >Sha | |||||
| AfterRipening.AUC | 011/415/111 | 35.8 | 16–25 | >Sha | ||||
| ABA.WS.Gmax | 011 | 5.4 | 12–34 | >Bay-0 | ||||
| ABA.WS.AUC | 011 | 4.0 | 16–37 | >Bay-0 | ||||
| Cold.NS.Fresh.Gmax | 7.4 | 12–30 | >Bay-0 | |||||
| Cold.NS.Fresh.AUC | 4.7 | 12–27 | >Bay-0 | |||||
| Heat.NS.Fresh.Gmax | 3.6 | 12–31 | >Sha | |||||
| Heat.WS.AR.Gmax | 011/415 | 8.1 | 17–37 | >Bay-0 | ||||
| Heat.WS.AR.AUC | 011/415 | 9.2 | 17–34 | >Bay-0 | ||||
| 5-1 | Stratification.AR.t50 | MSAT514 | 4.0 | 26.6/7.50 | 21–37 | >Bay-0 | ||
| NaCl.NS.t50 | 8.8 | 19–29 | >Bay-0 | |||||
| NaCl.NS.t10 | 214 | 11.3 | 21–30 | >Bay-0 | ||||
| NaCl.WS.t50 | 361 | 9.9 | 20–30 | >Bay-0 | ||||
| NaCl.WS.U8416 | 3.5 | 17.7–30 | >Bay-0 | |||||
| NaCl.WS.t10 | 5.4 | 18–30 | >Bay-0 | |||||
| CD.NS.AR.Gmax | 4.7 | 20–39 | >Bay-0 | |||||
| 5-2 | Stratification.Fresh.Gmax | 152 | MSAT59 | 4.6 | 57.8/17.25 | 48–68 | >Sha | Dormancy QTL; delay of germination, DOG1 (Bentsink et al., 2010) |
| Stratification.Fresh.AUC | 152 | 6.0 | 48–67 | >Sha | ||||
| AfterRipening.AR.Gmax | 152 | 7.7 | 51–67 | >Sha | ||||
| AfterRipening.AR.AUC | 152 | 11.7 | 50–62 | >Sha | ||||
| NaCl.NS.t50 | 4.3 | 52–70 | >Sha | QTL for cold-dark germination in Bay-0 × Sha, CDG-6 (Meng et al., 2008) | ||||
| Cold.NS.Fresh.U8416 | 1.3 | 47–61 | >Bay-0 | |||||
| Heat.NS.AR.AUC | 152 | 5.1 | 46–62 | >Sha | ||||
| Heat.WS.AR.AUC | 4.8 | 53–70 | >Sha | |||||
| 5-3 | NaCl.NS.Gmax | MSAT520037 | 10.1 | 67.4/20.04 | 61–72 | >Sha | QTL for germination on salt in Bay-0 × Sha, SALT SENSITIVITY RESPONSE2 (Vallejo et al., 2010) | |
| NaCl.NS.t10 | 6.4 | 62–79 | >Sha | |||||
| NaCl.NS.AUC | 11.6 | 58–71.6 | >Sha | |||||
| NaCl.WS.Gmax | 410/152 | 18.0 | 63–71.6 | >Sha | ||||
| NaCl.WS.t50 | 410/152/103 | 19.6 | 64–72 | >Sha | QTL for germination on salt in Sha × Columbia (Galpaz and Reymond, 2010) | |||
| NaCl.WS.U8416 | 5.4 | 59–74 | >Sha | |||||
| NaCl.WS.t10 | 18.4 | 65–74 | >Sha | |||||
| NaCl.WS.AUC | 23.2 | 64–73 | >Sha | |||||
| Mannitol.NS.Gmax | 152 | 13.2 | 63–74 | >Sha | ||||
| Mannitol.NS.AUC | 152 | 10.4 | 60–74 | >Sha | ||||
| Mannitol.WS.t50 | 152 | 4.0 | 63–77 | >Sha | ||||
| Mannitol.WS.AUC | 152 | 3.5 | 62.3–79 | >Sha | ||||
| Heat.NS.Fresh.Gmax | 4.8 | 55–73 | >Sha | |||||
| Heat.WS.AR.t50 | 152 | 8.0 | 63–74 | >Sha | ||||
| 5-4 | Stratification.Fresh.Gmax | 152 | MSAT519 | 3.9 | 85/25.92 | 80–91.2 | >Sha | Cloned mucilage-affecting locus, MUM2 (Macquet et al., 2007) |
| Stratification.Fresh.t50 | 10.4 | 79.1–91 | >Sha | |||||
| Stratification.Fresh.t10 | 16.3 | 79.1–88 | >Sha | |||||
| Stratification.Fresh.AUC | 152 | 7.3 | 80–91.2 | >Sha | ||||
| AfterRipening.t50 | 14.8 | 79.1–90 | >Sha | |||||
| AfterRipening.t10 | 19.3 | 79.1–88 | >Sha | |||||
| AfterRipening.AUC | 152 | 4.7 | 80–91.2 | >Sha | ||||
| Mannitol.NS.t50 | 9.6 | 80–91 | >Sha | |||||
| Mannitol.NS.U8416 | 5.1 | 80–91.2 | >Sha | |||||
| Mannitol.NS.t10 | 6.2 | 79.1–91 | >Sha | |||||
| ABA.NS.Gmax | 103 | 9.9 | 79–89 | >Bay-0 | ||||
| ABA.NS.t50 | 103 | 22.8 | 80–89 | >Bay-0 | ||||
| ABA.NS.t10 | 9.7 | 80–90 | >Bay-0 | |||||
| ABA.NS.AUC | 103 | 12.0 | 73–91 | >Bay-0 | ||||
| ABA.WS.Gmax | 410/152/103 | 8.2 | 81–90 | >Bay-0 | ||||
| ABA.WS.t50 | 103 | 19.5 | 79.1–88 | >Bay-0 | ||||
| ABA.WS.U8416 | 103 | 11.9 | 76–88 | >Bay-0 | ||||
| ABA.WS.AUC | 410/152/103 | 22.3 | 80–88 | >Bay-0 | ||||
| Heat.NS.AR.Gmax | 152 | 11.0 | 80–90 | >Sha | ||||
| Heat.NS.AR.AUC | 152 | 9.1 | 82–91 | >Sha | ||||
| Heat.WS.AR.U8416 | 103 | 13.3 | 80–91.2 | >Sha | ||||
| Size.imbibed.seed.Area | 103 | 72.3 | 79.1–89 | >Sha |
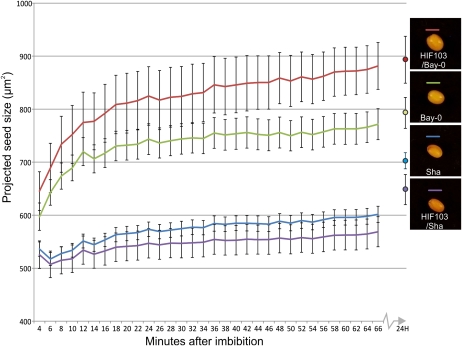
We detected a vast QTL for imbibed seed size at the bottom of chromosome V, which could be confirmed by the use of HIF103. Upon imbibition, seeds swell due to rapid water uptake and possibly because of the expansion of the inner mucilage layer. In Sha, which is a natural mutant for the mucilage-modified2 (mum2) gene (Macquet et al., 2007), this swelling did not occur. Also, the HIF lines at the mum2 position showed a clear difference in swelling phenotype, which was still significant 24 h after imbibition (Fig. 8).
Figure 8.
Different increases in seed size during the start of imbibition for Bay-0 (green), Sha (blue), HIF103/Bay-0 (red), and HIF103/Sha (purple) seeds. Shown is the average projected seed size area of 10 seeds. Error bars represent se values. Photographs show 24-h imbibed seeds.
DISCUSSION
When analyzing large (RIL) populations, it is hardly feasible to manually count all germination experiments several times a day to obtain germination curves. Therefore, previous studies were mostly restricted to counting end-point germination (Quesada et al., 2002; Alonso-Blanco et al., 2003; Clerkx et al., 2004; Laserna et al., 2008; Meng et al., 2008; Bentsink et al., 2010; Galpaz and Reymond, 2010; Vallejo et al., 2010). A germination curve allows QTL mapping under conditions where rate and uniformity are delayed but maximum germination is not affected. Therefore, we used the Germinator package (Joosen et al., 2010), which enables the measurement of cumulative germination data and the extraction of five germination parameters that describe the resulting germination curve. In this study, we describe several germination QTLs that were not detected before in the Bay-0 × Sha population. We observed interesting colocalizations for several germination traits and identified the loci that show large-effect epistatic interactions. Among these were new loci and loci similar to the ones already found in other RIL populations, as summarized in Table IV for the major identified QTL loci.
Dormancy
Primary dormancy has been studied extensively in various RIL populations (Bentsink et al., 2010). These authors quantified primary dormancy with the DSDS50 parameter (days of dry storage to reach 50% germination), which is a good measure for after-ripening-related dormancy breaking. Although we only compared the germination characteristics of freshly harvested seeds with those of after-ripened seeds and fresh seeds with and without stratification, we detected large genetic variation. Both dormancy-breaking treatments showed strong QTLs at positions 3-2, 4-1, and 5-2, colocating with DELAY OF GERMINATION6 (DOG6), DOG18, and DOG1, respectively (Table IV). DOG18 was not detected in a Landsberg erecta (Ler) × Sha population and showed a stronger dormancy in Ler as compared with Antwerp-1 (An-1), Feira-0 (Fei-0), and Kashmir-2 (Kas-2) (Bentsink et al., 2010). We detected stronger dormancy in Sha as compared with Bay-0 at the DOG18 locus. This suggests that both Ler and Sha contain an allele of similar strength that is stronger when compared with An-1, Fei-0, Kas-2, and Bay-0.
Remarkably, for both the DOG6 and DOG18 locations, the sensitivity to ABA was higher in Bay-0, whereas dormancy was deeper in Sha, which resulted in a directional change of the QTL effect. The more dormant Sha parent contains higher initial ABA levels (Supplemental Table S8), and apparently, after-ripening and stratification reduce the ABA sensitivity to a greater extent as compared with the Bay-0 parent. This effect was not observed for the DOG1 locus. Furthermore, we identified a strong effect of the dormancy-breaking treatments on the initiation (t10) and rate (t50) of germination at the bottom of chromosome V (marker MSAT519; 85 cM). The same was observed for germination on mannitol and germination at higher temperature. A QTL with an opposite effect at this position was found for germination on ABA. Interestingly, these colocated with a QTL found for imbibed seed size.
Water Uptake
Initiation and rate of germination are highly influenced by the overall water potential of the seed. The mucilage layer surrounding the seed appears to play an important role in the process of water uptake (Penfield et al., 2001). Sha is a natural mucilage mutant due to a mutation in the MUM2 gene, which changes the hydrophilic potential of rhamnogalacturonan I (Macquet et al., 2007). Although mucilage has been reported to be dispensable for germination and development under laboratory conditions (Arsovski et al., 2010), a link with germination under reduced water potential conditions was shown by Penfield et al. (2001). They showed reduced maximum germination of a mucilage-impaired mutant only on osmotic polyethylene glycol solutions. In our study, other traits that colocated on the MUM2 locus were delayed initiation and rate of germination on osmotic mannitol solution but also on water, which clearly shows the advantage of determining a detailed germination curve. We also observed a very strong QTL for swelling of the seed in the first hours of imbibition (imbibed seed size) at the MUM2 location. Interestingly, exogenous ABA can be used to stimulate mucilage production, and ABA deficient1 (aba1) mutants are affected in mucilage production (Karssen et al., 1983). This indicates a regulatory role of ABA in mucilage production and fits with our observation of the colocalization of a QTL for initiation and rate of germination with a QTL with the opposite effect for ABA sensitivity. Therefore, we hypothesize that Sha has a slower initiation and rate of germination, combined with reduced ABA sensitivity due to its mutation in the MUM2 gene. This observation may open new research strategies to define the regulatory role of ABA in mucilage production and its multiple effects on germination parameters.
Germination Responses to Salt, Heat, and ABA
At the top of chromosome I, underlying marker F12M12, we detected a strong QTL for maximum germination in the presence of 100 mm NaCl or 0.5 μm ABA. A similar locus has been identified and fine-mapped in a Ler × Sha population (Ren et al., 2010). They identified a premature stop codon in the Response to ABA and Salt1 gene (RAS1; At1g09950) in Sha that led to a truncated protein and showed its role as a negative regulator of salt tolerance during seed germination and early seedling growth by enhancing ABA sensitivity. Here, we show that a similar locus also is responsible for tolerance to germination at 30°C. This suggests an additional role for the RAS1 gene. Increased heat tolerance due to a modulation of ABA sensitivity has been shown before for other loci (Argyris et al., 2008; Lee et al., 2010). Interestingly, this study showed a strong effect of stratification that resulted in a strong reduction of significant linkage for NaCl, heat, and ABA sensitivity at the F12M12 locus. A specific QTL for germination on NaCl preceded by a cold stratification period was found at the middle of chromosome I (marker T27K12). Also at this locus, we found colocation with sensitivity for germination on ABA after stratification. Further fine-mapping at this locus might help to elucidate the effect of stratification on ABA-mediated abiotic stress tolerance as well as the apparent overlap of dormancy and stress responses.
Especially interesting is QTL 5-1 (Table IV; Fig. 6), which mainly influences rate and initiation of germination. We detected this QTL for t50 in after-ripened seeds with stratification treatment, but also for t10 and t50 for germination on salt, regardless of a preceding cold stratification, and for maximum germination after an accelerated aging treatment. One of the genes underlying this QTL interval is a nicotinamidase gene (NIC2; At5g23230), the mutant of which has retarded germination and impaired germination potential (Hunt et al., 2007). These authors suggested that NIC2 is normally metabolizing nicotinamide during moist chilling or after-ripening, which relieves the inhibition of poly(ADP-ribose) polymerase activity and allows DNA repair to occur prior to germination. Both accelerated aging and germination under salt-stress conditions might require optimal functioning of this DNA repair mechanism. Further research is needed to determine whether NIC2 is causal for this QTL.
Detection of epistatic interactions in genetic studies can enhance the understanding of underlying molecular mechanisms. Recently, Galpaz and Reymond (2010) showed strong epistasis in the genetic network controlling germination under salt stress in Arabidopsis. Due to careful dissection of the epistatic relationships, they were able to show that three detected QTLs rely on the presence of a Columbia allele at a QTL on the top of chromosome I. This observation led to the hypothesis that RAS1 (Ren et al., 2010) functions as a switch of the genetic network by regulating the expression of the other QTLs. In another study, it was found that epistasis significantly influences both fitness and germination in Arabidopsis (Huang et al., 2010), and novel allele combinations were identified that resulted in higher fitness. In our study, we detected clear hotspots of epistatic interactions between QTL on chromosomes III, IV, and V (ATHCHIB2 + MSAT332, MSAT435, and MSAT520037 + MSAT519, respectively). This observation strengthens the hypothesis that some of the traits with strong QTL colocalizations indeed rely on the same underlying genetic networks.
CONCLUSION
In conclusion, we analyzed natural variation for many seed germination characteristics and showed their correlation, (shared) QTL positions, and epistatic interactions using a high-throughput phenotyping approach and subsequent high-throughput QTL mapping. Using the HIF approach, confirmation of some major QTL hotspots was demonstrated, which allows a fast but solid confirmation of a QTL position. Together with results from several other studies focusing on genetic variation in seed traits, this study has generated an extensive QTL database for Arabidopsis and proposed a method of analysis to visualize the genetic landscape of seed performance. This database is a solid resource for further study. For most of the loci found in this and other studies, further characterization, and in most cases fine-mapping, must be undertaken to elucidate the causal molecular mechanisms. Furthermore, we have designed a free available analysis protocol to perform detailed high-throughput QTL analysis based on the R/qtl MQM routine. In this era of large-scale phenotyping, we regard a detailed analysis of QTL, QTL×QTL, and QTL×E interaction as indispensable steps to allow the visualization and interpretation of multiple traits. Finally, there is great potential in combining extensive phenotyping of RIL populations with available “omics” approaches to increase the speed of causal allele detection (Joosen et al., 2009).
MATERIALS AND METHODS
Plant Growth
Seeds from the core population (165 lines) of an Arabidopsis (Arabidopsis thaliana) Bay-0 × Sha recombinant inbred population (Loudet et al., 2002) were obtained from the Versailles Biological Resource Centre for Arabidopsis (http://dbsgap.versailles.inra.fr/vnat/). The population is mapped with 69 markers with an average distance between the markers of 6.1 cM (Loudet et al., 2002). Maternal plants were grown twice in a fully randomized setup. In the first round, we separated the harvest into three groups (A, B, and C), each containing three to five plants per RIL. In the second round, we pooled the harvest of four to seven plants per RIL (D). Plants were grown on 4- × 4-cm rockwool plugs (MM40/40; Grodan) and watered with 1 g L−1 Hyponex fertilizer (nitrogen:phosphorus:potassium, 7:6:19; http://www.hyponex.co.jp) in a climate chamber (20°C day, 18°C night) with 16 h of light (35 W m−2) at a relative humidity of 70%. Seeds were bulk harvested and after-ripened at room temperature and ambient relative humidity until they reached their maximum germination potential after 5 d of imbibition.
Seed Germination Assays
Germination experiments were performed as described previously (Joosen et al., 2010). Germination was scored using the Germinator package. When mentioned, a cold stratification period of 4 d at 4°C in the dark was applied before transferring the trays to the germination incubator (20°C, continuous light). A temperature of 10°C was used for testing “cold” germination, whereas 30°C was used for “heat” germination. Salt stress was applied by replacing the water by a NaCl (Sigma-Aldrich; S-3014) solution (100 mm for nonstratified seeds and 125 mm for stratified seeds, as stratification reduces the sensitivity to NaCl). A solution of −0.6 MPa mannitol (Sigma-Aldrich; 15719) was used to test for osmotic stress. ABA (Duchefa Biochemie; A0941) was initially dissolved in a few drops of 1 n NaOH from which stock solutions were prepared in 10 mm MES buffer, pH 5.9. ABA was used at a final concentration of 0.5 μm. Accelerated aging of the seeds was performed using a closed container with a saturated NaCl solution to obtain 75% relative humidity. Seeds were equilibrated for 7 d in this humidity at 20°C in the dark, followed by 30 d at 75% relative humidity and 32.5°C in the dark (Hundertmark et al., 2011). For each measurement, we used at least two replicates for every harvest that was tested (Table I) to determine the germination characteristics. All germination tests were performed in a fully randomized setup. Averages were calculated and corrected for their proper control (Table I). For Gmax and AUC, the stress condition was subtracted from the control condition. Because t10, t50, and U8416 are reversed parameters, we subtracted control conditions from stress conditions. Dry seed size was determined by taking closeup photographs from approximately 100 to 200 seeds using a Nikon D80 camera with a 50-mm Macro objective. Imbibed seed size was extracted from the first images acquired within the Germinator setup (100–200 seeds). For Figure 8, 10 seeds of each line were photographed with maximum magnification (using a 50-mm Macro objective). The photographs were analyzed using the open-source image-analysis suite ImageJ (http://rsbweb.nih.gov/ij/) by using color thresholds combined with particle analysis.
Single-Trait QTL Mapping Using R/qtl
This protocol is provided as an R script and performs the following analysis and transformations; steps specified with an (o) are optional, and (c) means that the steps can be configured by the user.
Preparing and Starting
To run an analysis, two kinds of files need to be provided: (1) the raw data file formatted in R/qtl cross format (see the manual of R/qtl for more information); and (2) a configuration file. The description of the allowed (and necessary) parameters is available in the manual (Supplemental File S3). After having prepared these files, the user can start R and change to the directory the script is located in by using the setwd command:
> setwd(“d:/script”)
Now the script can be loaded by using the source command:
> source(“qtl_analysis_script.R”)
Then, the analysis is started by the doAnalysis command; this command takes two parameters: a filename and a directory (only needed when different from the script directory):
> doAnalysis(“myconfig.txt”,”d:/configfiles”)
The script will now start the analysis.
Analysis Protocol: Loading and Preprocessing
The script starts by reading the configuration and data file and then performs the following analysis:
(o,c) Use a Z-transformation to check data distribution and remove any outliers.
(o,c) Automated phenotype normalization/user-supplied normalization.
(c) Plotting of basic genetic statistics, such as genetic map, recombination frequencies, and trait-correlation plots.
Main Analysis Loop (Performed for All Traits)
In the main loop, we analyze traits independently. For each trait, a directory in the output folder is created to store the per-trait results. We used the following steps.
Plotting of basic per-phenotype statistics: distributions of raw and normalized data:
(c) QTL modeling using backward elimination on genetic cofactors, followed by interval mapping using multiple cofactors (multiple QTL mapping).
(o) QTL by single-marker mapping using Haley-Knott regression (scan.one).
(o) Whole-genome interaction scan heat map made by using the scan.two QTL interaction-mapping routine from R/qtl.
(o) Single-trait permutation using the MQM permutation routine.
Generation of various plots related to the single-trait QTL results:
Raw phenotype effect plots.
QTL model by backward elimination.
QTL profiles showing scan.one, scan.two, and the MQM interval-mapping results.
Interaction effect plots.
Furthermore, at the end of the analysis for each phenotype, additional information is saved. Also, an R data file containing the output object from the MQM scan is saved to enable the user to cancel the analysis and resume at a later time.
Multiple-Trait Analysis and Plots
After all phenotypes have been analyzed, the script provides additional plots based on the aggregated data.
(o,c) Circle plots showing selected cofactors and possible interactions.
(o,c) Combined heat maps and Hclust clustering of all QTL profiles.
(o,c) Extraction of the clusters and plotting of the grouped QTL results.
(o) The user can output .sif-formatted files, which can be used to create network overviews in Cytoscape (or other visualization tools). We provide two networks:
The QTL network: genetic marker network based on QTL data.
The epistatic interaction network produced by summarizing the interactions found between selected cofactors from the MQM algorithm.
To visualize the created .sif files, download and install Cytoscape, launch Cytoscape, and load the network using File/Import/Network (multiple file types).
QTL×E Analysis
By using Genstat version 14, the variance-covariance model is calculated for the G+GxE variation in the phenotypic data based on an unstructured model. Given this variance-covariance model, a simple interval mapping procedure was started followed by two rounds of composite interval mapping using detected QTLs as cofactors but omitting these covariables in windows around the simple interval mapping QTLs. A final multiple-QTL model is created using a backward elimination for significant cofactors. For imputing virtual markers along the chromosomes, a step size of 2 cM was used. Minimum cofactor proximity as well as minimum separation for selected QTLs were set to 16 cM.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. LOD score correlation plot comparing raw and transformed data (fitted to a normal distribution).
Supplemental Table S1. Pearson correlation matrix for AUC trait values measured in four different harvests, Microsoft Excel (xls) format.
Supplemental Table S2. Cross-object containing all data for the 327 measured traits (in columns) in the 165 RILs (in rows), which can be used as input file for the script, comma-separated value (csv) format.
Supplemental Table S3. Cross-object containing data for the average values of the 94 traits (in columns) in the 165 RILs (in rows) that are described in this paper.
Supplemental Table S4. Output file that summarizes all QTL results, providing an overview of all detected QTLs, their peak LOD scores, positions, confidence intervals, and directions for all 327 traits measured, Microsoft Excel (xls) format.
Supplemental Table S5. Output file that summarizes all QTL results, providing an overview of all detected QTLs, their peak LOD scores, positions, confidence intervals, and directions for all 94 traits described in this paper, Microsoft Excel (xls) format.
Supplemental Table S6. Genstat version 14 output summary for effect size and explained variance for QTL×E effects, Microsoft Excel (xls) format.
Supplemental Table S7. Overview of t test P values for all contrasts from the HIF results.
Supplemental Table S8. Results and methodology from ABA measurement in dry Bay-0 and Sha seeds.
Supplemental File S1. QTLnetwork.cys: Cytoscape file containing the interactive QTL network shown in Figure 5.
Supplemental File S2. Interactionnetwork.cys: Cytoscape file containing the epistatic interaction network shown in Figure 7.
Supplemental File S3. Package.zip: zip file containing all necessary files for reanalysis of the presented data, including a manual with installation and analysis instructions.
Supplementary Material
Acknowledgments
We thank Suzanne Abrams and Irina Zaharia from the Plant Hormone Profiling Laboratory (National Research Council Plant Biotechnology Institute) for carrying out hormone measurements (http://www.nrc-cnrc.gc.ca/eng/facilities/pbi/plant-hormone.html). We also thank Martin Boer (Department of Biometris, Wageningen University and Research Centre) and Linus van der Plas (Laboratory of Plant Physiology, Wageningen University) for useful comments and critical reading of the manuscript.
References
- Alonso-Blanco C, Aarts MG, Bentsink L, Keurentjes JJ, Reymond M, Vreugdenhil D, Koornneef M. (2009) What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21: 1877–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Bentsink L, Hanhart CJ, Blankestijn-de Vries H, Koornneef M. (2003) Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 164: 711–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arends D, Prins P, Jansen RC, Broman KW. (2010) R/qtl: high-throughput multiple QTL mapping. Bioinformatics 26: 2990–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyris J, Dahal P, Hayashi E, Still DW, Bradford KJ. (2008) Genetic variation for lettuce seed thermoinhibition is associated with temperature-sensitive expression of abscisic acid, gibberellin, and ethylene biosynthesis, metabolism, and response genes. Plant Physiol 148: 926–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsovski AA, Haughn GW, Western TL. (2010) Seed coat mucilage cells of Arabidopsis thaliana as a model for plant cell wall research. Plant Signal Behav 5: 796–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriere Y, Laperche A, Barrot L, Aurel G, Briand M, Jouanin L. (2005) QTL analysis of lignification and cell wall digestibility in the Bay-0 × Shahdara RIL progeny of Arabidopsis thaliana as a model system for forage plant. Plant Sci 168: 1235–1245 [Google Scholar]
- Bentsink L, Hanson J, Hanhart CJ, Blankestijn-de Vries H, Coltrane C, Keizer P, El-Lithy M, Alonso-Blanco C, de Andrés MT, Reymond M, et al. (2010) Natural variation for seed dormancy in Arabidopsis is regulated by additive genetic and molecular pathways. Proc Natl Acad Sci USA 107: 4264–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer MP, Wright D, Feng L, Podlich DW, Luo L, Cooper M, van Eeuwijk FA. (2007) A mixed-model quantitative trait loci (QTL) analysis for multiple-environment trial data using environmental covariables for QTL-by-environment interactions, with an example in maize. Genetics 177: 1801–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling R, Li Y, Tesson BM, Fu J, Wu C, Wiltshire T, Gerrits A, Bystrykh LV, de Haan G, Su AI, Jansen RC. (2008) Genetical genomics: spotlight on QTL hotspots. PLoS Genet 4: e1000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890 [DOI] [PubMed] [Google Scholar]
- Calenge F, Saliba-Colombani V, Mahieu S, Loudet O, Daniel-Vedele F, Krapp A. (2006) Natural variation for carbohydrate content in Arabidopsis: interaction with complex traits dissected by quantitative genetics. Plant Physiol 141: 1630–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang GC, Barua D, Kramer EM, Amasino RM, Donohue K. (2009) Major flowering time gene, flowering locus C, regulates seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA 106: 11661–11666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Schumaker K, Zhu JK. (2004) Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J Exp Bot 55: 225–236 [DOI] [PubMed] [Google Scholar]
- Clerkx EJM, El-Lithy ME, Vierling E, Ruys GJ, Blankestijn-De Vries H, Groot SPC, Vreugdenhil D, Koornneef M. (2004) Analysis of natural allelic variation of Arabidopsis seed germination and seed longevity traits between the accessions Landsberg erecta and Shahdara, using a new recombinant inbred line population. Plant Physiol 135: 432–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechaine JM, Gardner G, Weinig C. (2009) Phytochromes differentially regulate seed germination responses to light quality and temperature cues during seed maturation. Plant Cell Environ 32: 1297–1309 [DOI] [PubMed] [Google Scholar]
- Diaz C, Saliba-Colombani V, Loudet O, Belluomo P, Moreau L, Daniel-Vedele F, Morot-Gaudry JF, Masclaux-Daubresse C. (2006) Leaf yellowing and anthocyanin accumulation are two genetically independent strategies in response to nitrogen limitation in Arabidopsis thaliana. Plant Cell Physiol 47: 74–83 [DOI] [PubMed] [Google Scholar]
- Eeuwijk FAV, Malosetti M, Boer MP. (2007) Modelling the genetic basis of response curves underlying genotype × environment interaction. Spiertz JHJ, Struik PC, Van Laar HH, , Scale and Complexity in Plant Systems Research: Gene-Plant-Crop Relations. Springer, Dordrecht, The Netherlands, pp 113–124 [Google Scholar]
- Elwell AL, Gronwall DS, Miller ND, Spalding EP, Brooks TL. (2011) Separating parental environment from seed size effects on next generation growth and development in Arabidopsis. Plant Cell Environ 34: 291–301 [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G. (2006) Seed dormancy and the control of germination. New Phytol 171: 501–523 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD. (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell (Suppl) 14: S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Keurentjes JJ, Bouwmeester H, America T, Verstappen FW, Ward JL, Beale MH, de Vos RC, Dijkstra M, Scheltema RA, et al. (2009) System-wide molecular evidence for phenotypic buffering in Arabidopsis. Nat Genet 41: 166–167 [DOI] [PubMed] [Google Scholar]
- Galpaz N, Reymond M. (2010) Natural variation in Arabidopsis thaliana revealed a genetic network controlling germination under salt stress. PLoS ONE 5: e15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutterman Y. (2000) Maternal effects on seeds during development. Fenner M, , The Ecology of Regeneration in Plant Communities, Ed 2. CABI, Wallingford, UK, pp 59–84 [Google Scholar]
- Haley CS, Knott SA. (1992) A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity (Edinb) 69: 315–324 [DOI] [PubMed] [Google Scholar]
- Huang X, Schmitt J, Dorn L, Griffith C, Effgen S, Takao S, Koornneef M, Donohue K. (2010) The earliest stages of adaptation in an experimental plant population: strong selection on QTLs for seed dormancy. Mol Ecol 19: 1335–1351 [DOI] [PubMed] [Google Scholar]
- Hundertmark M, Buitink J, Leprince O, Hincha DK. (2011) The reduction of seed-specific dehydrins reduces seed longevity in Arabidopsis thaliana. Seed Sci Res 21: 165–173 [Google Scholar]
- Hunt L, Holdsworth MJ, Gray JE. (2007) Nicotinamidase activity is important for germination. Plant J 51: 341–351 [DOI] [PubMed] [Google Scholar]
- Jansen RC. (1993) Interval mapping of multiple quantitative trait loci. Genetics 135: 205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RC. (2008) Quantitative trait loci in inbred lines. Handbook of Statistical Genetics. John Wiley & Sons, Chichester, UK, pp 587–622 [Google Scholar]
- Jansen RC, Van Ooijen JW, Stam P, Lister C, Dean C. (1994) Genotype-by-environment interaction in genetic mapping of multiple quantitative trait loci. Theor Appl Genet 91: 33–37 [DOI] [PubMed] [Google Scholar]
- Joosen RV, Kodde J, Willems LA, Ligterink W, van der Plas LH, Hilhorst HW. (2010) GERMINATOR: a software package for high-throughput scoring and curve fitting of Arabidopsis seed germination. Plant J 62: 148–159 [DOI] [PubMed] [Google Scholar]
- Joosen RV, Ligterink W, Hilhorst HW, Keurentjes JJ. (2009) Advances in genetical genomics of plants. Curr Genomics 10: 540–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karssen CM, Brinkhorst-van der Swan DLC, Breekland AE, Koornneef M. (1983) Induction of dormancy during seed development by endogenous abscisic acid: studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh. Planta 157: 158–165 [DOI] [PubMed] [Google Scholar]
- Laserna MP, Sanchez RA, Botto JF. (2008) Light-related loci controlling seed germination in Ler × Cvi and Bay-0 × Sha recombinant inbred-line populations of Arabidopsis thaliana. Ann Bot (Lond) 102: 631–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Kang JY, Park HJ, Kim MD, Bae MS, Choi HI, Kim SY. (2010) DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiol 153: 716–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tesson BM, Churchill GA, Jansen RC. (2010) Critical reasoning on causal inference in genome-wide linkage and association studies. Trends Genet 26: 493–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkies A, Müller K, Morris K, Turecková V, Wenk M, Cadman CS, Corbineau F, Strnad M, Lynn JR, Finch-Savage WE, et al. (2009) Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: a comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell 21: 3803–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F. (2002) Bay-0 × Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet 104: 1173–1184 [DOI] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Krapp A, Daniel-Vedele F. (2003a) Quantitative trait loci analysis of water and anion contents in interaction with nitrogen availability in Arabidopsis thaliana. Genetics 163: 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Merigout P, Talbotec J, Daniel-Vedele F. (2003b) Quantitative trait loci analysis of nitrogen use efficiency in Arabidopsis. Plant Physiol 131: 345–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Gaudon V, Trubuil A, Daniel-Vedele F. (2005) Quantitative trait loci controlling root growth and architecture in Arabidopsis thaliana confirmed by heterogeneous inbred family. Theor Appl Genet 110: 742–753 [DOI] [PubMed] [Google Scholar]
- Loudet O, Michael TP, Burger BT, Le Metté C, Mockler TC, Weigel D, Chory J. (2008) A zinc knuckle protein that negatively controls morning-specific growth in Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 17193–17198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Saliba-Colombani V, Camilleri C, Calenge F, Gaudon V, Koprivova A, North KA, Kopriva S, Daniel-Vedele F. (2007) Natural variation for sulfate content in Arabidopsis thaliana is highly controlled by APR2. Nat Genet 39: 896–900 [DOI] [PubMed] [Google Scholar]
- Macquet A, Ralet MC, Loudet O, Kronenberger J, Mouille G, Marion-Poll A, North HM. (2007) A naturally occurring mutation in an Arabidopsis accession affects a β-D-galactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage. Plant Cell 19: 3990–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malosetti M, Voltas J, Romagosa I, Ullrich SE, van Eeuwijk FA. (2004) Mixed models including environmental covariables for studying QTL by environment interaction. Euphytica 137: 139–145 [Google Scholar]
- Meng P-H, Macquet A, Loudet O, Marion-Poll A, North HM. (2008) Analysis of natural allelic variation controlling Arabidopsis thaliana seed germinability in response to cold and dark: identification of three major quantitative trait loci. Mol Plant 1: 145–154 [DOI] [PubMed] [Google Scholar]
- Moreau L, Charcosset A, Gallais A. (2004) Use of trial clustering to study QTL × environment effects for grain yield and related traits in maize. Theor Appl Genet 110: 92–105 [DOI] [PubMed] [Google Scholar]
- Murtagh F. (1985) A survey of algorithms for contiguity-constrained clustering and related problems. Comput J 28: 82–88 [Google Scholar]
- Orsi CH, Tanksley SD. (2009) Natural variation in an ABC transporter gene associated with seed size evolution in tomato species. PLoS Genet 5: e1000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne RW, Harding SA, Murray DA, Soutar DM, Baird BD. (2011) The Guide to GenStat Release 14. Part 2. Statistics. VSN International, Hemel Hempstead, UK [Google Scholar]
- Penfield S, Meissner RC, Shoue DA, Carpita NC, Bevan MW. (2001) MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 13: 2777–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, García-Martínez S, Piqueras P, Ponce MR, Micol JL. (2002) Genetic architecture of NaCl tolerance in Arabidopsis. Plant Physiol 130: 951–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Zheng Z, Chinnusamy V, Zhu J, Cui X, Iida K, Zhu JK. (2010) RAS1, a quantitative trait locus for salt tolerance and ABA sensitivity in Arabidopsis. Proc Natl Acad Sci USA 107: 5669–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond M, Svistoonoff S, Loudet O, Nussaume L, Desnos T. (2006) Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ 29: 115–125 [DOI] [PubMed] [Google Scholar]
- Rivero-Lepinckas L, Crist D, Scholl R. (2006) Growth of plants and preservation of seeds. Methods Mol Biol 323: 3–12 [DOI] [PubMed] [Google Scholar]
- Tamura N, Yoshida T, Tanaka A, Sasaki R, Bando A, Toh S, Lepiniec L, Kawakami N. (2006) Isolation and characterization of high temperature-resistant germination mutants of Arabidopsis thaliana. Plant Cell Physiol 47: 1081–1094 [DOI] [PubMed] [Google Scholar]
- Tuinstra MR, Ejeta G, Goldsbrough PB. (1997) Heterogeneous inbred family (HIF) analysis: a method for developing near-isogenic lines that differ at quantitative trait loci. Theor Appl Genet 95: 1005–1011 [Google Scholar]
- Vallejo AJ, Yanovsky MJ, Botto JF. (2010) Germination variation in Arabidopsis thaliana accessions under moderate osmotic and salt stresses. Ann Bot (Lond) 106: 833–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK. (2002) Cell signaling during cold, drought, and salt stress. Plant Cell (Suppl) 14: S165–S183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.