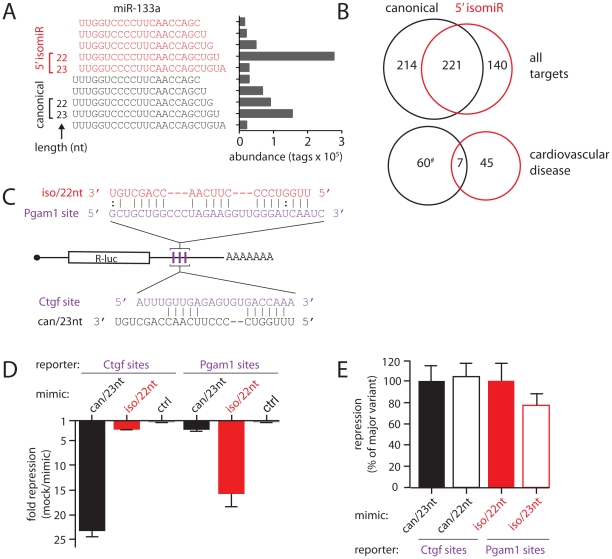
Figure 3. isomiRs of miR-133a with different targeting properties.
(A) Major mature miR-133a species and their abundance in HL-1 cardiomyocytes. Sequence tags are grouped into those with canonical (black, ‘can’) and +1 (red, ‘iso’) 5′ start sites. Brackets denote sequences used as miRNA mimics in panels D and E. (B) Venn diagrams indicate number and overlap of predicted mRNA targets (Targetscan) for canonical and +1 5′ isomiR variants of miR-133a (top: all predicted targets, bottom: only targets with roles in cardiovascular disease (Ingenuity; #: significant enrichment of gene function term, p<0.01). (C) Schematic of reporter constructs made to contain three copies of predicted miR-133a binding sites from Pgam1 or Ctgf mRNA 3′UTRs. Base pairing potential between sites and miR-133a isomiRs is also shown. (D) Ctgf or Pgam1 R-luc reporters were transfected into HeLa cells with mimics of the major variants of the canonical (can/23 nt) and +1 5′ variants (iso/22 nt) of miR-133a, an irrelevant control, or no mimic at all, and luciferase activity measured 24 hours later. The fold change of expression is calculated as no mimic/mimic and results are averages of four independent experiments with standard error. (E) Transfections as in (D) comparing mimics of different lengths (can/23 nt vs can/22 nt, iso/22 nt vs. iso/23 nt; for sequence see panel A). Repression is given as a percentage of that seen with the respective major variant and results are averages of at least three independent experiments with standard error.