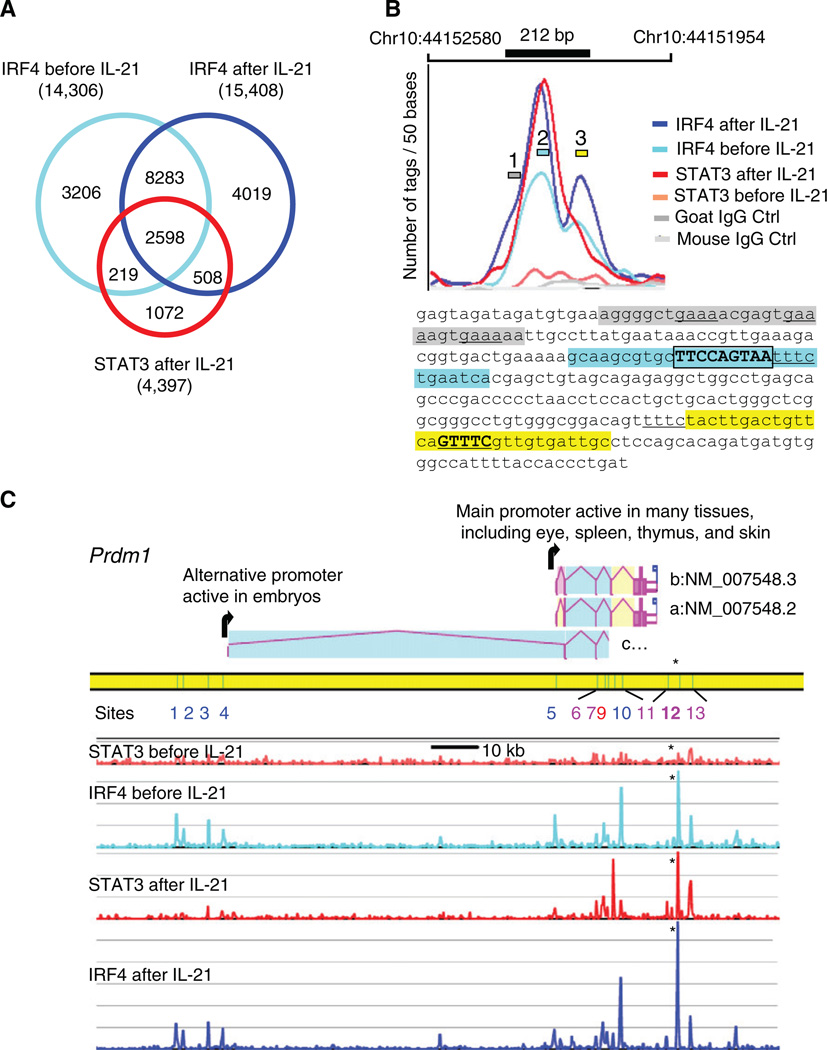
Figure 5. STAT3- and IRF4-Binding Sites Colocalize.
(A) Venn diagram of IRF4-binding sites before and after IL-21 stimulation. Shown are STAT3 sites after stimulation; the rare STAT3 sites before IL-21 treatment were omitted.
(B) High-resolution mapping of STAT3 and IRF4 ChIP-Seq tags at the Prdm1 IL-21 response element. The number of tags/50 bp is plotted. The 212 bp IL-21 response element is drawn to scale and exactly overlaps the STAT3 and two IRF4-binding sites. The maxima of the curves coincide with the essential motifs shown in Figures 3 and 4; the sequences at these maxima are high-lighted in blue and yellow, with DNA-binding motifs for STAT3 and IRF4 boxed and underlined, respectively. The shoulder of the largest IRF4 peak suggests a secondary binding site (gray region).
(C) The mouse Prdm1 gene includes three alternative transcript variants a, b, and c (AceView at NCBI). The STAT3- and IRF4-binding sites before and after IL-21 treatment are aligned (scale bar is 10 kb and 20 tags/200 bp window vertically), with binding sites corresponding to peaks 1–13 (Table S11). Although only one STAT3-IRF4 site (site 12, see asterisk, shown in detail in B) was found to regulate IL-21-mediated Prdm1 expression, several other major STAT3 peaks were induced by IL-21, and all overlapped IRF4 peaks. In intron 5 of variant a, site 9 strongly binds STAT3 but weakly binds IRF4, whereas site 10, which is 1.59 kb 3′ of site 9, strongly binds IRF4 but weakly binds STAT3. Additionally, IRF4 associated to the two promoter areas at sites 1 to 5.