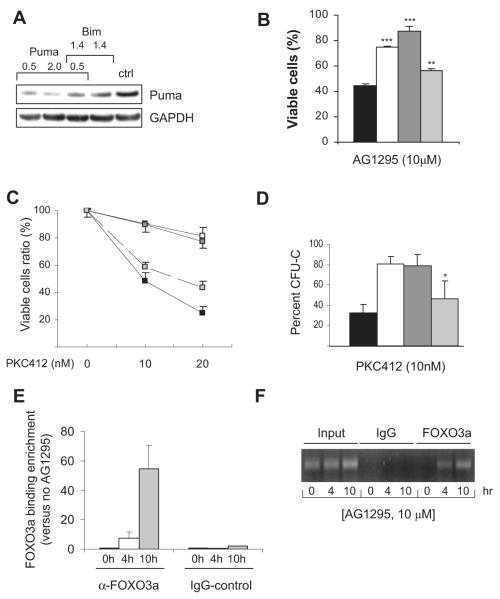
Figure 7. Bim is more critical than Puma in apoptotic induction of FLT3-ITD inhibition and is a direct transcriptional target of FOXO3a in FLT3-ITD cells treated with AG1295.
(A,B) FDC-P1/FLT3-ITD cells were transfected with siRNA control pmaxGFP (■) or siRNA to Bim (□), Bim + Puma ( ), or Puma (
), or Puma ( ). Cell lysates were prepared after 24 hours for Western blot analysis of Puma (A). In separate experiments, cells were transfected with siRNA as indicated, and 4 hours after transfection, AG1295 at 10 μM was added. Twenty-four hours later, cells were analyzed for apoptosis by flow cytometry after staining with annexin V–FITC and PI (B). Data shown are mean (± SD) from 1 representative experiment performed in duplicate and repeated twice. **P < .01 (SD over control siRNA); ***P < .001 (SD over control siRNA). (C,D) Wild-type (■), bim-/- (□), bim-/- puma-/- (
). Cell lysates were prepared after 24 hours for Western blot analysis of Puma (A). In separate experiments, cells were transfected with siRNA as indicated, and 4 hours after transfection, AG1295 at 10 μM was added. Twenty-four hours later, cells were analyzed for apoptosis by flow cytometry after staining with annexin V–FITC and PI (B). Data shown are mean (± SD) from 1 representative experiment performed in duplicate and repeated twice. **P < .01 (SD over control siRNA); ***P < .001 (SD over control siRNA). (C,D) Wild-type (■), bim-/- (□), bim-/- puma-/- ( ), or puma-/-(
), or puma-/-( ) bone marrow–derived Lin− progenitor cells infected with FLT3-ITD were FACS-sorted based on expression of EYFP. After treatment with 10 and 20 nM PKC412, the viability was assessed by flow cytometry after 72 hours in culture and compared with cells cultured without treatment (C). The bone marrow cells were also analyzed for colony formation in the absence of supportive cytokine but treated with 10 nM PKC412. Colony numbers were assessed after 7 days of culture (D). Data are mean (± SD) from 3 experiments. *P < .03 (SD compared with bone marrow–derived colonies from bim-/- mice). (E,F) ChIP-quantitative PCR analysis for FOXO3a binding to the Bim promotor. Sonicated DNA from FDC-P1/FLT3-ITD cells treated with AG1295 at 10 μM for 4 and 10 hours was immunoprecipitated with anti-FOXO3a or control rabbit IgG and amplified by quantitative PCR using primers specific for the Bim promotor. Relative expression of Bim was normalized to the input value and then compared with the corresponding untreated samples. Error bars represent SEM of triplicate reactions from 1 representative analysis of 2 separate experiments performed (E). PCRs were also visualized on 1.5% agarose gels stained with ethidium bromide (F).
) bone marrow–derived Lin− progenitor cells infected with FLT3-ITD were FACS-sorted based on expression of EYFP. After treatment with 10 and 20 nM PKC412, the viability was assessed by flow cytometry after 72 hours in culture and compared with cells cultured without treatment (C). The bone marrow cells were also analyzed for colony formation in the absence of supportive cytokine but treated with 10 nM PKC412. Colony numbers were assessed after 7 days of culture (D). Data are mean (± SD) from 3 experiments. *P < .03 (SD compared with bone marrow–derived colonies from bim-/- mice). (E,F) ChIP-quantitative PCR analysis for FOXO3a binding to the Bim promotor. Sonicated DNA from FDC-P1/FLT3-ITD cells treated with AG1295 at 10 μM for 4 and 10 hours was immunoprecipitated with anti-FOXO3a or control rabbit IgG and amplified by quantitative PCR using primers specific for the Bim promotor. Relative expression of Bim was normalized to the input value and then compared with the corresponding untreated samples. Error bars represent SEM of triplicate reactions from 1 representative analysis of 2 separate experiments performed (E). PCRs were also visualized on 1.5% agarose gels stained with ethidium bromide (F).