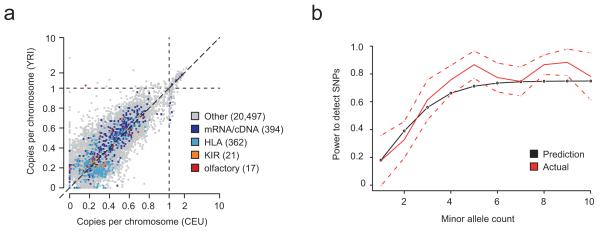
Figure 4.
Population analysis with Cortex. (a) Estimates of mean copy number per genome in CEU and YRI for novel sequence contigs identified from analysis of pooled population graphs for 164 humans sequenced to low depth (2-4x) with the 1000 Genomes Project. Contigs are all at least 100bp long and have <90% homology to the reference genome (determined by BLAST). Allelic variation lies in the interval (0,1), while copy-number variable sequence can be present up to 6.3 times. Variants are annotated by whether they show significant homology to known transcripts, including HLA, KIR and olfactory receptor (OR) genes as specific categories, which are clearly enriched. We note the presence of several OR matches that are approximately 20% frequency in YRI but apparently absent from CEU. (b) Power to detect SNP variants previously analysed through SNP genotyping in HTS data from 10 chimpanzees (6x coverage, 50bp reads analysed at k=31). Empirical estimates (red, with normal approximation to binomial confidence intervals shown dotted) closely track predictions (black) from the theoretical model.