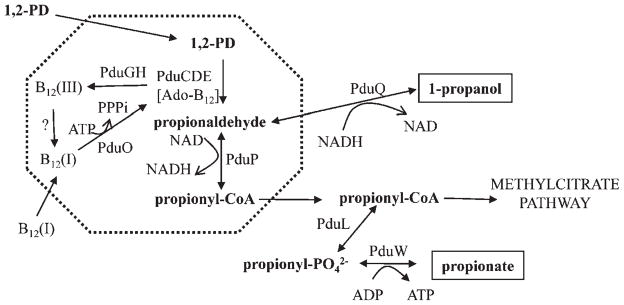
Figure 3.
Model for 1,2-propanediol degradation by S. enterica. The dashed line indicates the shell of the microcompartment which is composed of seven BMC-domain proteins (PduABB′JKTU). The first two steps of 1,2-propanediol degradation (conversion of 1,2-propanediol to propionyl-CoA) occur in the lumen of the microcompartment and the remaining steps in the cytoplasm. The proposed function of this microcompartment is sequestration of propionaldehyde to minimize its toxicity. The Ado-B12 cofactor is sometimes converted to B12(III) in a catalytic by-reaction inactivating diol dehydratase. B12(III) is released from diol dehydratase (PduCDE) by a reactivase (PduGH), and reduced to B12(I) by an unknown enzyme. Then, ATP:cob(I)alamin adenosyltransferase (PduO) converts B12(I) to active cofactor (Ado-B12) which associateswith diol dehydratase to form active holoenzyme. Abbreviations: 1,2-PD, 1,2-propanediol; PduCDE, coenzyme B12-dependent diol dehydratase; PduP, propionaldehyde dehydrogenase; PduL, phosphotransacylase; PduW, propionate kinase; PduQ, 1-propanol dehydrogenase.