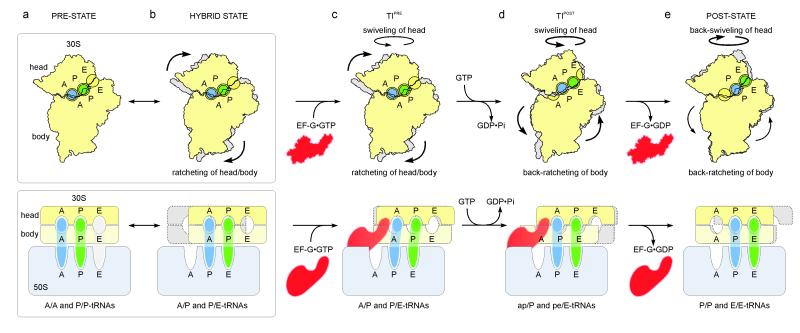
Fig. 4. Model for translocation.
a, b, The PRE ribosome exists in a dynamic equilibrium between (a) base states with classical A/A- and P/P-tRNAs and (b) rotated states with hybrid A/P- and P/E-tRNAs6,7,9-11. c, Binding of EF-G•GTP to (a) PRE- or (b) hybrid-states stabilizes the ratcheted state12 as observed in the TIPRE. d, Fast GTP hydrolysis by EF-G17 accelerates translocation via an unlocking step on the 30S subunit18. Domain IV of EF-G uncouples un-ratcheting from the reverse movement of the A/P- and P/E-tRNAs back into classical states i.e. a doorstop function. Through head-swiveling and un-ratcheting motion, the tRNAs move from aa/P and pp/E into the 30S intra-subunit ap/P and pe/E hybrid states. e, Complete un-ratcheting of the 30S subunit leads to the POST state 70S•EF-G complex19. Back-swiveling of the 30S-head re-establishes tRNAs in the classical (pp/P) P- and E- (ee/E) states. Translocation is completed by dissociation of EF-G•GDP.