Figure 4.
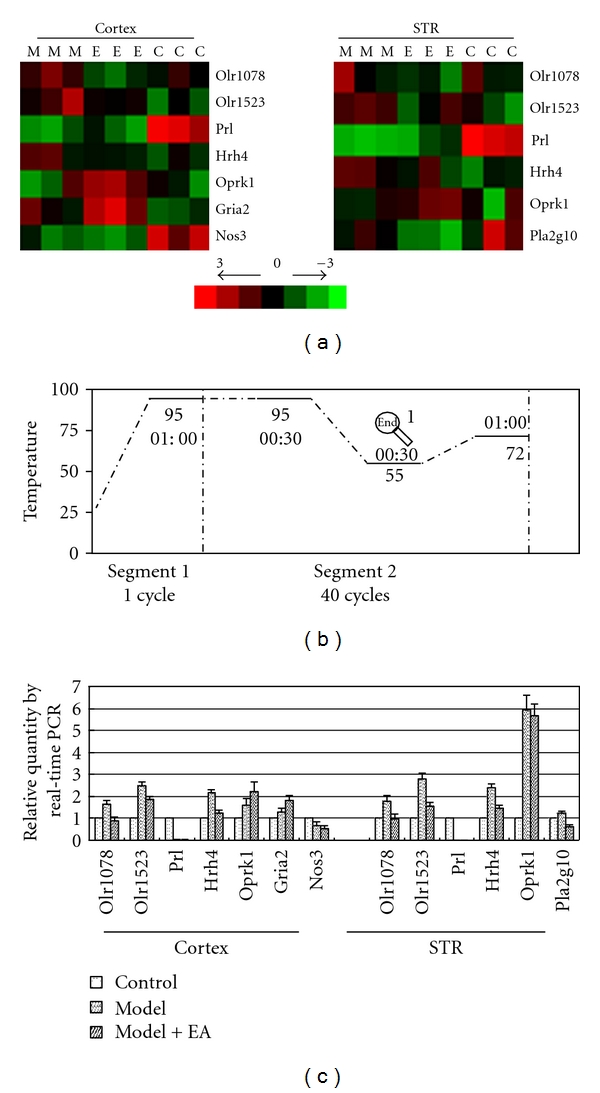
Selected transcripts clustered and validated with real-time PCR. After pathway analysis, we selected genes involved in major pathways (the common pathways in the cortex and STR, Table 7). First, selected genes were clustered using Cluster 3.0 according to the LOG value in the STR groups (a) Red is relatively upregulated and green is relatively downregulated in different samples. The three “Cs” are the three control samples. The three “Ms” are the model samples. The three “Es” are the EA-treated samples. Then, to verify the reliability of the microarray analysis, we verified these selected genes from the clustering diagram using real-time RT-PCR. The real-time PCR primers are listed on Table S1 (devised and synthesized by Takara). The reaction procedure was present on (b). If the level of control was regarded as 1, the corresponding gene expression in model and EA-treated groups were present on (c). These transcripts analyzed here showed coherent profiles with cluster (a).
