Fig 6.
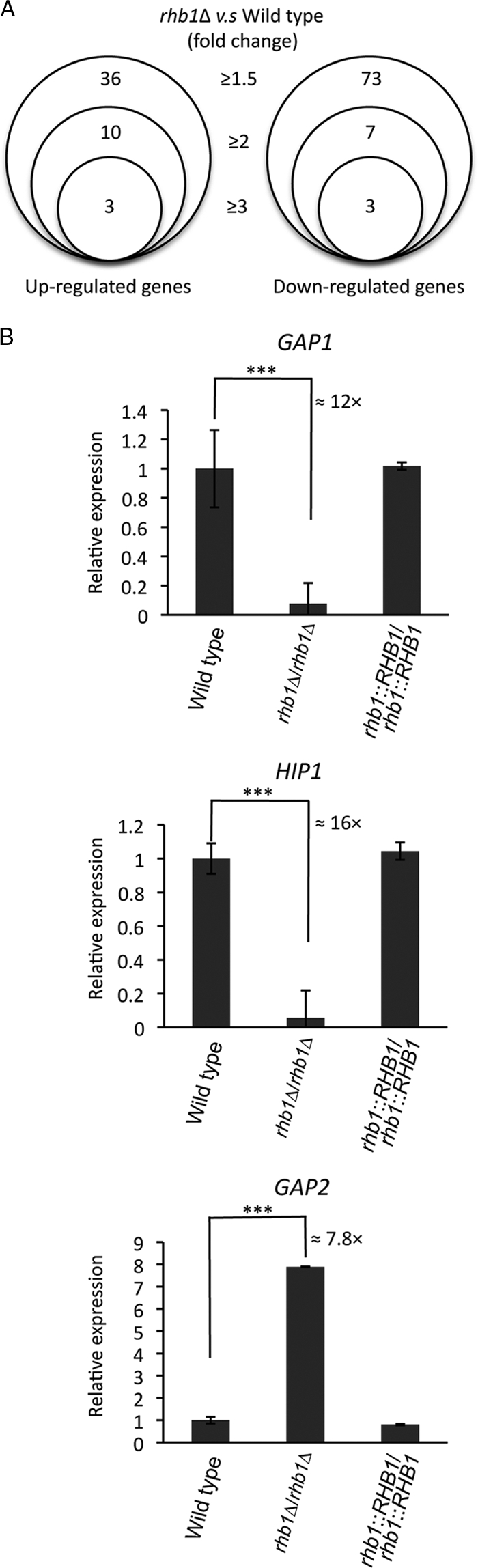
Identification of Rhb1 target genes by the use of DNA microarray analysis. (A) Summary of the fold change thresholds of the differentially expressed genes in the rhb1-deleted mutant (CCT-D1) compared with the wild type (SC5314). RNAs were collected after both strains were grown to mid-log phase in SD medium at 30°C. (B) Relative expression levels of three general amino acid permease genes: GAP1, HIP1, and GAP2. Quantitative real-time PCR was performed for GAP1, HIP1, and GAP2 in the wild-type (SC5314), CCT-D1 (rhb1Δ/rhb1Δ), and CCT-RD1 (RHB1-reconstituted) strains. Cells were grown in SD medium for 4 h to mid-log phase under the same conditions used in the microarray experiment. The threshold cycle (CT) value of each gene was derived from the average of the results of 3 experiments. The ΔCT value was determined by subtracting the average ΔCT of endogenous control EFB1 from the average CT of the gene tested. The ΔΔCT of each gene was calculated by subtracting the ΔCT value of the corresponding calibration value (wild-type sample). The average ΔΔCT and standard deviation values were determined from experiments performed in triplicate. The relative expression level of each gene is 2−ΔΔCT. ***, P ≤ 0.005. The error bars show the standard deviation.
