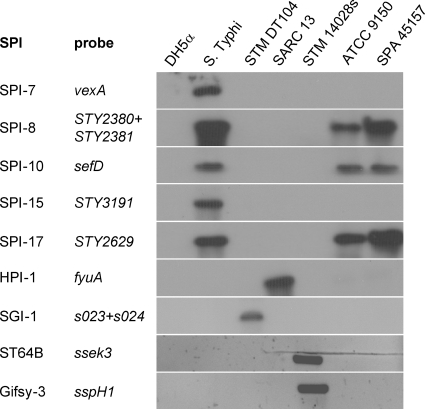
Fig 2.
Molecular typing of the S. Paratyphi A outbreak strain. Southern blot hybridization and PCR were used to determine the presence of the Salmonella pathogenicity islands 7, 8, 10, 15, and 17, HPI, SGI-1, and the virulence-associated prophages ST64B and Gifsy-3 in the genome of the outbreak strain (SPA 45157). E. coli DH5α (DH5α) was used as a negative control for all analyses and S. Typhi CT18 (S. Typhi), S. Typhimurium DT104 (STM DT104), Salmonella Reference Collection C 13 (SARC-13), and S. Typhimurium 14028s (STM 14028s) were used as positive controls. The specific genes used as probes to determine the presence of the above loci are listed. The presence of the prophages ST64B and Gifsy-3 was detected by PCR using primers specific to sseK3 and sspH1, respectively, while the other images show the results of a Southern blot hybridization using the nonradioactive DIG system.