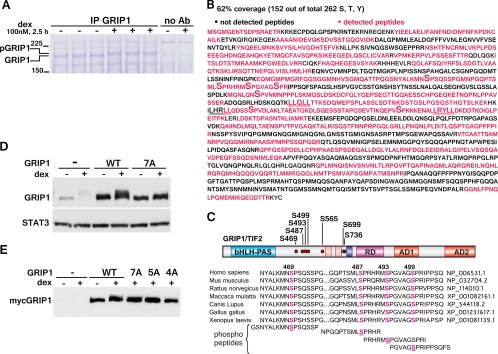
Fig 3.
The identification of GRIP1 phosphorylation sites by MS. (A) U2OS-rGR cells (∼8 × 108 cells/condition) were treated with or without dex, and GRIP1 was immunoprecipitated with 50 μg GRIP1 antibody/condition. After separation on SDS-PAGE gel, the proteins were visualized by Coomassie blue staining. Ab, antibody. (B) MS analysis of GRIP1. Percent coverage was calculated from the results of two independent experiments, and the MS-detected peptides are shown in magenta. LXXLL motifs are underlined; phosphorylated serines are marked by a large “S.” (C) GRIP1 phosphosites identified by MS. The GRIP1 diagram depicts the bHLH-PAS domain, NR boxes, RD, and AD 1 and 2. Six dex-inducible (red dots) and one constitutive (blue dot) phosphorylation site are marked. The alignment of the GRIP1 S469/S487/S493/S499 phosphosite cluster between different species and the peptides used for raising phosphosite-specific antibodies are shown. (D, E) MS-identified phosphosites mediate the GRIP1 mobility shift. myc-tagged WT GRIP1 or the 7A (all identified sites), 5A (S469/487/493/499/565A), or 4A (S469/487/493/499A) mutant was transfected into U2OS-rGR cells (250 ng of GRIP1 plasmid/250,000 cells, overnight), which were then untreated or treated with dex for 2 h and harvested, and GRIP1 electrophoretic mobility was evaluated by immunoblotting with antibodies to GRIP1 (D) and the myc tag (E).